Hola todos,
I currently have several .csv tables from a statistical collection, each with different measured variables but at least one identifier variable in common. Each identifier refers to a company for example.
My goal is to merge all these independent excel tables in order to obtain a final data.frame with X columns of variables and one and only one row per observation.
If I manage to succeed, it will be extremely simple to perform all the statistical operations I want to do on it.
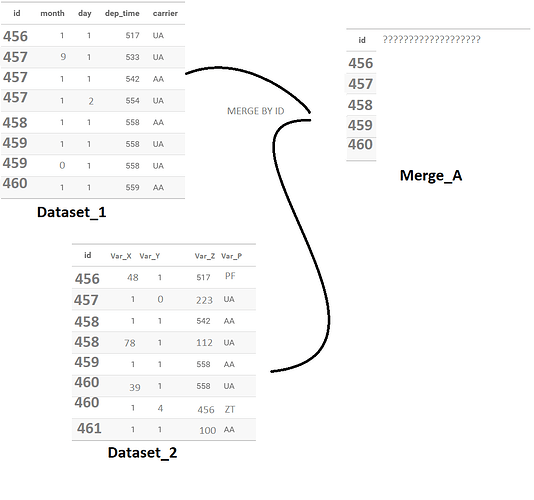
The problem is unfortunately the following, there are several observations that share the same identifier, which obviously made sense when I collected the data (which I did not do) to distinguish the activity sites within the same company
The problem is that I have to do my analysis on these data and that this necessarily poses a problem when I try to merge the datasets with merge.
Here's some reproducible code to explain it:
# Packages - - - -
library(dplyr)
library(tidyverse)
# Load data - - - -
Dataset_1 <- read.csv("XXXXXXXX")
Dataset_2 <- read.csv("XXXXXXXX")
Dataset_3 <- read.csv("XXXXXXXX")
Dataset_4 <- read.csv("XXXXXXXX")
# Code - - - -
# I merge by iteration, dataframe by dataframe but I ignore if it's possible to do both dataframe in one time
Merge_A<-merge(Dataset_1, Dataset_2,
by=0,all.x = T,all.y = T)
Merge_B<-merge(Merge_A,Dataset_3,
by=0,all.x = T,all.y = T)
Final_data<-merge(Merge_B,Dataset_4,
by=0,all.x = T,all.y = T)
# And here normally I've got my perfect data to save
And by explain it with pictures :
I try to make it as clear as possible, this is the method I personally imagined in order to be able to get out of it but maybe much simpler solutions exist in reality. Maybe the solution is actually obvious, but my brain is starting to boil and I'm not used to dealing with this kind of problem unfortunately.
Hope you will be able to help me !