Hi R-Community,
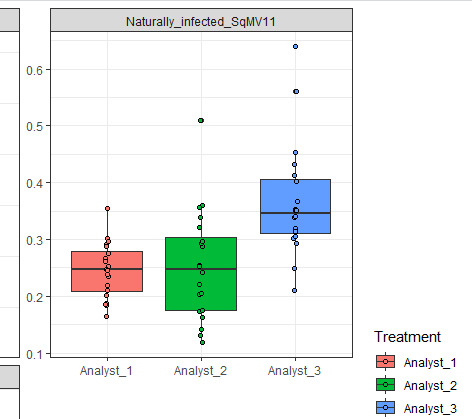
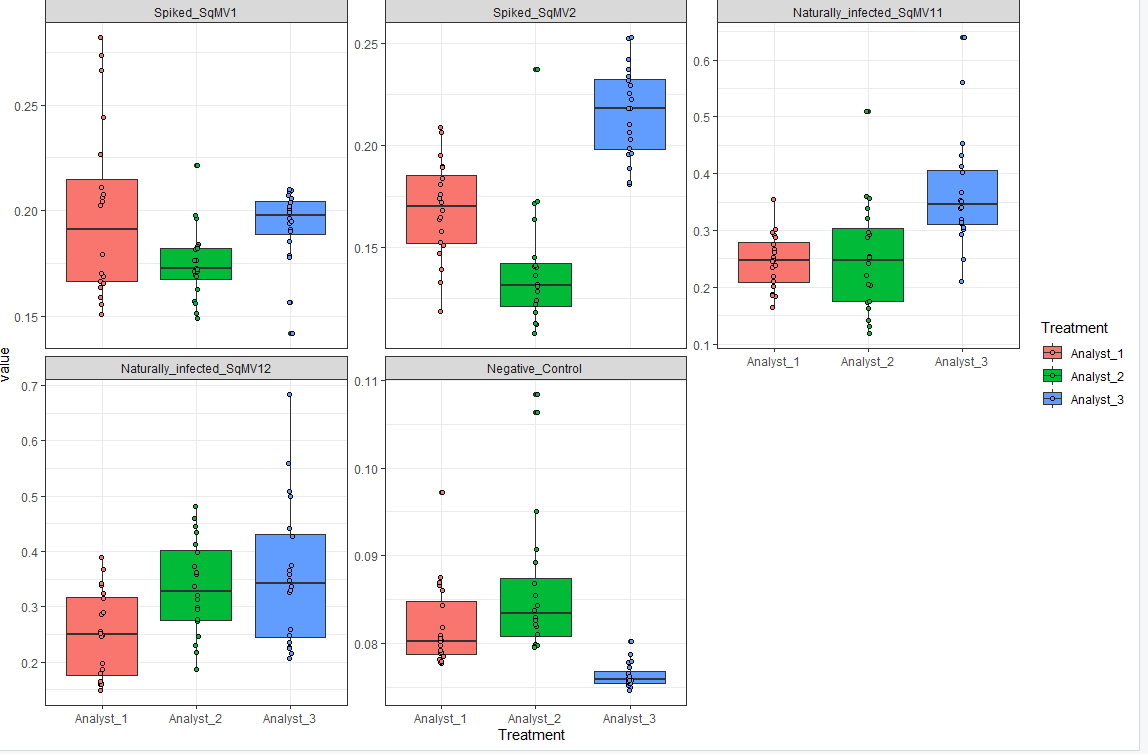
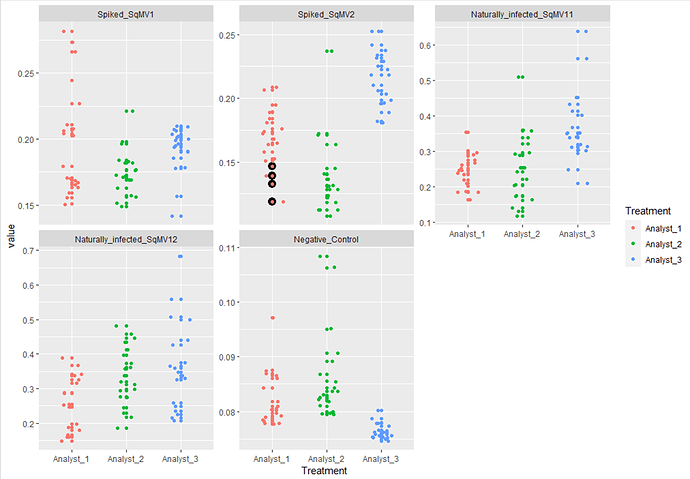
I plotted a boxplot using ggplot2 library as attached below. Now, I am interested in highlighting specific data values with in the treatment. How can I accomplish this? For example, I want to change the color of specific data point for analyst_2 and different specific data point for analyst_1.
Any help will be highly appreciated!
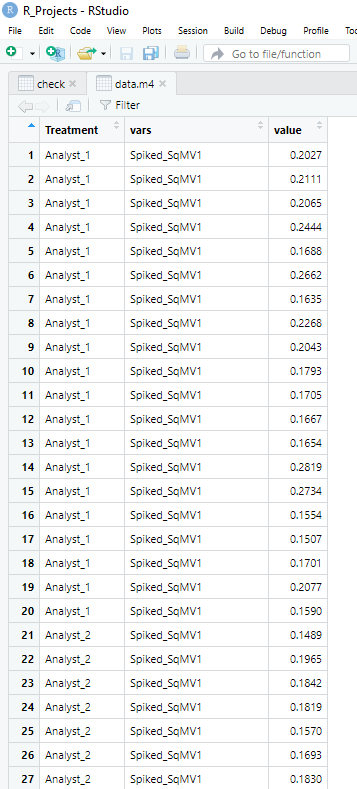
I code I use to generate the plot is below:
ggplot(data.m4, aes(x = Treatment, y = value, fill = Treatment)) + geom_boxplot(position = position_dodge(width = 0.7)) + facet_wrap(~vars, scales = "free_y") + geom_point(position = position_jitterdodge(jitter.width = 0.1, dodge.width = 0.7), aes(fill = Treatment), pch = 21) + theme_bw()