This may be a trivial question, but when using MonetDBLite, are you able to run other packages using the same R objects? For example, I'm imagining have a table that is too large to fit in Memory, and doing modeling using the caret package (or perhaps some other packages that are more niche).
If i understood your question correctly, you want to have the data in a Monetdb instance and then run functions from the caret package on it, right?
If this is the case then I believe this is not possible because the caret functions need to be somehow translated to the db backend. The functions that MonetDB understands "just like that" are listed on the dplyr db backend vignette + on the MonetDB docs on their website.
At least not with the Lite version of MonetDB - see this discussion for example.
But there is the "normal" standalone MonetDB which does support in-database running of R UDFs (user-defined functions). There are some nice examples on their website of how to do this and much more.
Hi @david2, I tried out MonetDB a couple of days ago. Even though there is no SQL translation written specifically for MonetDB in either package, you can get it to work by pointing the translation to the MySQL translation: https://github.com/tidyverse/dplyr/issues/8#issuecomment-344773402
To you broader point, you're correct regarding the ability to run models (caret) inside MonetDB, or any database.
Ah I see, so it would seem that if one were to use these options, there would be restrictions and likely the need to adapt workflows specifically to use the available tools that MonetDB et al can accommodate. Unfortunately in my case it is quite essential to be able to run models and various other UDF so may need to invest in some cloud space.
Hey Edgar,
Cool that you tried it out - however i'm not sure if we are talking about the same package here because there are actually 2 - MonetDB.R and MonetDBLite.
I'm a bit confused by your comment
Even though there is no SQL translation written specifically for MonetDB in either package...
In the comment on the issue, I guess you mean there is no translation for the MonetDB.R pkg - however there is also the MonetDBLite pkg which works fine with the whole suite.
My understanding is that the former (which offers finer control) has been repackaged into the latter (nicer format but less control).
Sorry if i'm misunderstanding or missing something obvious - i'm not that familiar with the dbplyr and the translation environments you mention in the comment.
You're right, we were talking about different packages. I need to give MonetDBLite a test drive!
ok cool. 
yeah, give it a go it's truly awesome software.
I have been looking for a replacement of MonetDBLite for my own workflow.
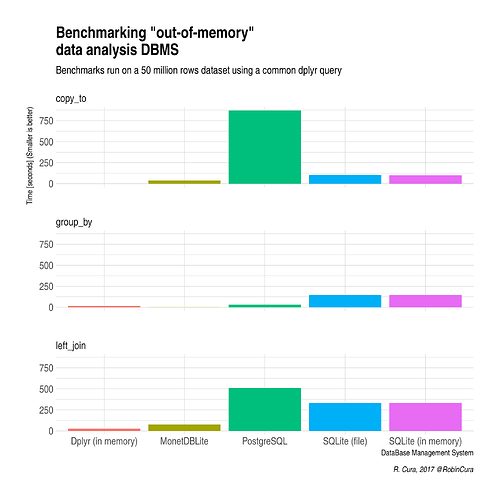
I'm working with ~ 50M rows databases, and was not very satisfied/reassured with MonetDBLite as I'm having some corruption in my DB from time to time.
Looking for an alternative, I did run some benchmarks that can be summed up in this plot :
So, for me, although I'm not that much confortable using MonetDBLite, I'll keep using it as there seems to be no free alternative for running "real-time" analysis (I'm making plots with shiny out of this DB) with such quantity of data.
Is there any way to update a MonetDBLite table using data from R? All of these examples don't discuss mutating or updating the tables. But that seems like a big part of data analysis. For my workflow, I would like to load a huge csv to a MonetDBLite instance, query tables, conduct analysis in R, like building models, and insert the new results into the table by adding columns. Is this not possible?
I know dbplyr does not mutate data on principle. But is there a generic way (using DBI) to update an entire MonetDBLite column or add a new one to an existing table? Especially without using paste!
yes, depending on what you want to do there are different routes you can take (although i'm kinda curious what you mean with paste 
- you can update a table in MonetDBLite by using the
dplyrbackend e.g.
library("tidyverse")
library("MonetDBLite")
library("DBI")
con <- dbConnect( MonetDBLite::MonetDBLite() , ":memory:" )
dbWriteTable(con, "mtcars", mtcars)
t <- tbl(con, "mtcars")
# add some new cols to t
t <-
t %>%
mutate(
new_disp = rank(disp),
mpg_vs_cyl = mpg / cyl,
cos_vs = cos(vs)
)
# write back mutated tbl into db
dbWriteTable(con, "mtcars", t, overwrite = TRUE)
For more details on what functions dplyr and MonetDBLite understand please check out my previous answer above and the links i provided there. Alternative route was suggested to you by Hannes in the github issue discussion you opened - that way is more powerful because the standalone MonetDB (note that this is different than the MonetDBLite pkg) understands arbitrary user-defined R functions.
hth.
Thank you for the response. Does the mutate occur in the database? It is my understanding that dplyr does not mutate the source data. But if dplyr only "stages" the mutation until I request it be explicitly written then that would work for my needs. I would like to avoid pulling the entire dataset into memory. Does this method do that?
The paste method I've seen in certain examples (not MonetDBLite) updated a database column by pasting an update query string with each value from R. This seemed really inefficient to me for large tables.
In the new version of dbplyr the copy_to() command is smart enough to know that you are using layz-dplyr code (t) and uses the resulting SQL, instead of downloading the data first, which guarantees that the mutate() happens inside the database: copy_to(con, t, "mtcars")
yes, as also Edgar pointed out this doesn't happen in main memory (in the sense that you do not pull all the data into R in order to compute this - there are other db-specific intricacies which are beyond this discussion).
Basically, thanks to dbplyr and dplyr, a certain subset of R's functions can be translated and executed on the db backend easily.
For a richer functionality and arbitrary R functions you need to take another route (e.g. standalone MonetDB).
I have recently found disk.frame package.
This package enables you to deal with larger-than-RAM data by both dplyr and data.table syntax. You can also use any R functions to manipulate data unlike other packages such as ff and bigmemory.
The approach of the package is to chunk large data into files via fst package and process them in parallel by future package.
It seems very promising package.
Oh wow! I am very happy someone found my package useful! I am almost ready to submit to CRAN!
This is what disk.frame is designed to solve! Give it a spin!
This is a harder problem and disk.frame can't solve that yet. But I have some experimental implementation where I connected disk.frame to biglm, speedglm, keras, and xgboost to develop some models. It's very experimental but I think it will work. Give it more time! I wish I can find a job where I can focus on this full-time and flesh it out!
I had a look at it now and its phenomenal! I will absolutely be using this starting even today. Its fantastic work - really well done and I hope you keep developing it!
I'll be keeping a very keen eye on this - I really look forward to seeing it work with xgboost/Keras. Again really well done!
And I see this is the gift that keeps on giving I see it even provides a way to convert SAS files to csv in chunks (!!!!)
Please PM me if you want to get the Sas conversion utility (a separate binary file which isn't included yet). It's at least 10 times faster than the nearest competitor in terms of speed.
But again the sas conversion code doesn't work without the SAS conversion utility.
This topic was automatically closed 21 days after the last reply. New replies are no longer allowed.
If you have a query related to it or one of the replies, start a new topic and refer back with a link.