alla
February 2, 2023, 12:03pm
1
Hello everyone!
So, I have created phyloseq object with next code:
taxonomy<-parse_taxonomy(taxonomy$data)
I got:
phyloseq
Now I can do bar plots, but I need to assign order for our samples.
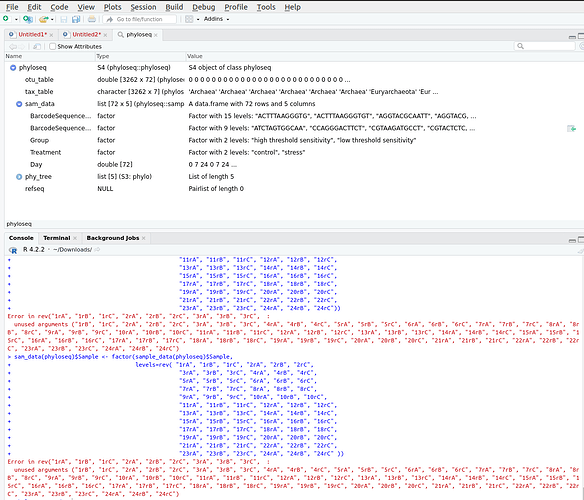
And i got this error:$<-.data.frame(S3Part(x, TRUE), name, value) :
How can I fix it?
I tried to reproduce your error; I could only do so by mis-stating the name from which the samples are ; so review that ...
library("phyloseq"); packageVersion("phyloseq")
data(soilrep)
sample_data(soilrep)
mylevels <- levels(sample_data(soilrep)$Sample)
#works
sample_data(soilrep)$Sample <- factor(sample_data(soilrep)$Sample ,
levels=rev(mylevels))
#gives the error
sample_data(soilrep)$SampleNOTRIGHTNAME <- factor(sample_data(soilrep)$SampleNOTRIGHTNAME ,
levels=rev(mylevels))
alla
February 2, 2023, 1:00pm
3
Thank you.
Error in rev(" some sample names ", :
rev is arbitrary; it reverses a list of values; in this case the old levels.
alla
February 2, 2023, 1:04pm
5
Oh, sorry for tat
Error in rev("1rA", "1rB", "1rC", "2rA", "2rB", "2rC", "3rA", "3rB", "3rC", :
sample id from metadata file I have 72 samples
alla
February 2, 2023, 1:08pm
6
Maybe this would be helpful
if you want to use rev; to reverse the order of your levels; if you are writing them out as a list by hand; you would need to use the c() function to collate them
rev(c( "1rA", "1rB", "1rC", "2rA", "2rB", "2rC", "3rA " # ... etc)
alla
February 2, 2023, 1:24pm
8
Thank you
Error in $<-.data.frame(S3Part(x, TRUE), name, value) :
ok; I think you are confused about what my code was about;
your screenshot does not show me anything called SampleID that you may have been trying to use.
alla
February 2, 2023, 1:37pm
10
Ok
alla
February 2, 2023, 1:42pm
11
Oh, what a pity.
lets walk real through the issue slowly.
names(sample_data(soilrep))
when you do the equivalent ; what are the names ?
names(sample_data(phyloseq))
alla
February 2, 2023, 2:08pm
13
Thank you)
names(sample_data(phyloseq))
I used all those objects to create the phyloseq object
head(metadata)#q2:types categorical categorical categorical categorical numeric
head(taxonomy)
head(features)
$type
$format
$contents
$version
$data
Also, maybe important
head(phyloseq)
so there is no SampleID present here to adjust; we agree ?
alla
February 2, 2023, 2:15pm
15
Sure)
Sorry I don't know; I'm not a phyloseq user and haven't studied their workflow.
vaulot
February 4, 2023, 7:21pm
17
A quick answer...
To do barplots 16S data, I do not recommend using phyloseq which is better for quick analysis. I usually export the data from phyloseq to data frames and plot the data frames using dplyr and ggplot2. It is much more flexible...
Also just having a quick look at your code, I will not use factors to recode the Samples names in PhyloSeq because in factors recode a set of values to 1, 2, 3 etc... and I am not sure Phyloseq will like that.
alla
February 5, 2023, 5:05pm
18
nirgrahamuk:
.
Thank you for your answer, but I don't use phyloseq to build barplots.
I used this tutorial
It worked, but the sample order is not what we need.
system
March 19, 2023, 5:06pm
19
This topic was automatically closed 42 days after the last reply. New replies are no longer allowed.