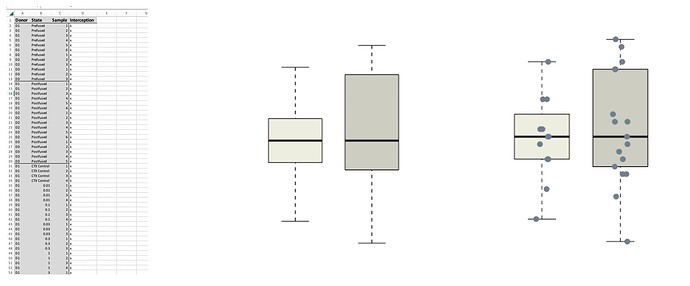
Hi! I've put this on my normal reddit forum but no help there so I thought id try here!
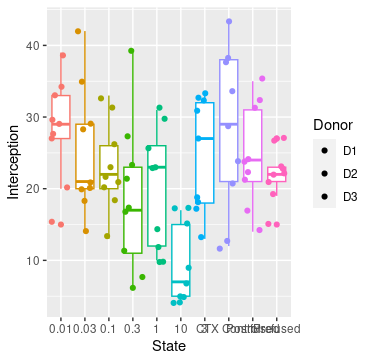
I have overlayed a stripchat onto my box & whisker, looks beautiful. But all the points are one colour when actually they're samples from 3 different donors: D1, D2 and D3 seperated in the .csv as a column called "Donor.
My Box and Whisker is Interception~State, two separate variables.
I have attached a picture of my excel (points replaced with x), a sample bar with and without the scatter.
My R Script reads:
boxplot (Interception~State, data=data.CTXScreen,
xlab="Sample State, including CTX treatment (µM)",
ylab = expression('Fibre Midline Interception'), main="Fibre Midline Interception for each state",
cex.main=2.5,
cex.lab=1.5,
cex.axis=1,
boxwex=0.7,
col= c('ivory2', 'ivory3', 'red1', 'darkorange', 'yellow', 'springgreen4', 'dodgerblue1', 'purple4', 'purple1', 'deeppink'))
stripchart(Interception~State, data=data.CTXScreen,
method = "jitter",
pch = 19,
col = c('slategrey'),
vertical = TRUE, add = TRUE)
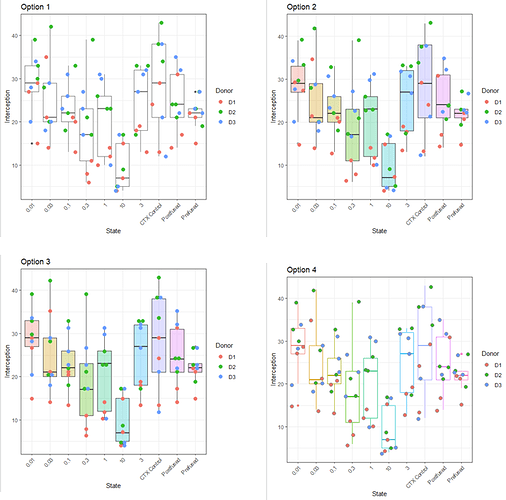
I've been messing around attempting things like this:
V2=runif(data.CTXScreen$Donor == "D1") {col = c('slategrey')}
else if (data.CTXScreen$Donor == "D2") {col = c('dark grey')}
else (data.CTXScreen$Donor == "D3") {col = c('black')}
x <- if(data.CTXScreen$Donor == "D1") {col = c('slategrey')}
y <- if (data.CTXScreen$Donor == "D2") {col = c('darkgrey')}
z <- if (data.CTXScreen$Donor == "D3") {col = c('black')}
df = data.frame(V1=runif(Donor == "D1", col = c('slategrey')), V2=runif(Donor == "D2", col = c('darkgrey')), V3=runif(Donor == "D3", col = c('black')))
df = data.frame(V1=sample.int(Donor == "D1", col = c('slategrey'), replace=TRUE), V2=runif(Donor == "D2", col = c('darkgrey')), V3=runif(Donor == "D3", col = c('black')))
Overall a very big mess and I wonder if anyone could give me a hand. I feel like its such a simple issue, but its had me stumped since about August!
This post is most helpful but I just can't see to translate it to my script