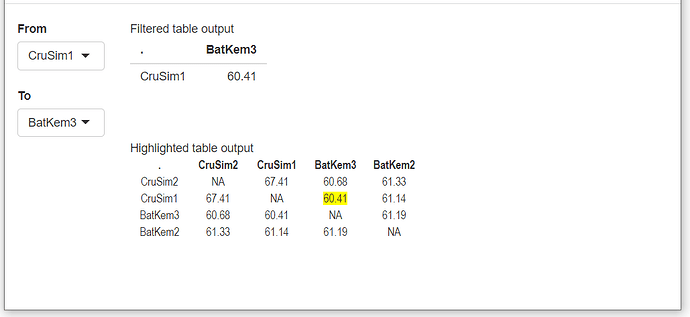
Hi,
I am a new Shiny user and I would like to create an app that allows a user to simultaneously filter rows and columns from a distance matrix. I guess the widget selectInput would alow me to make choices to select from the matrix below
| | CruSim2 | CruSim1 | BatKem3 | BatKem2 | ValMicro2 | BatKem1 | BatSten1_a | BatSten1_b | VallMicro1 | MagHoe1 | Amag_GK30097_PI468340_UGA_B | Ahoeh_VP9094_PI688954_Embrapa_B |
|----------------------------------|---------|---------|---------|---------|-----------|---------|------------|------------|------------|---------|-----------------------------|---------------------------------|
| CruSim2 | | 67.41 | 60.68 | 61.33 | 60.12 | 60.58 | 61.04 | 59.68 | 56.52 | 56.90 | 49.66 | 51.03 |
| CruSim1 | 67.41 | | 60.41 | 61.14 | 60.14 | 60.38 | 60.95 | 59.66 | 55.84 | 56.28 | 49.64 | 51.10 |
| BatKem3 | 60.68 | 60.41 | | 61.19 | 60.56 | 61.53 | 61.80 | 61.92 | 54.96 | 55.24 | 49.07 | 50.95 |
| BatKem2 | 61.33 | 61.14 | 61.19 | | 65.15 | 65.56 | 62.16 | 60.46 | 55.02 | 55.53 | 48.57 | 50.49 |
| ValMicro2 | 60.12 | 60.14 | 60.56 | 65.15 | | 67.64 | 61.81 | 59.89 | 54.93 | 54.85 | 48.50 | 50.42 |
| BatKem1 | 60.58 | 60.38 | 61.53 | 65.56 | 67.64 | | 62.19 | 60.80 | 54.63 | 54.93 | 48.62 | 50.60 |
| BatSten1_a | 61.04 | 60.95 | 61.80 | 62.16 | 61.81 | 62.19 | | 65.63 | 54.90 | 55.22 | 48.38 | 50.05 |
| BatSten1_b | 59.68 | 59.66 | 61.92 | 60.46 | 59.89 | 60.80 | 65.63 | | 54.22 | 54.34 | 48.42 | 50.40 |
| VallMicro1 | 56.52 | 55.84 | 54.96 | 55.02 | 54.93 | 54.63 | 54.90 | 54.22 | | 69.09 | 53.80 | 48.30 |
| MagHoe1 | 56.90 | 56.28 | 55.24 | 55.53 | 54.85 | 54.93 | 55.22 | 54.34 | 69.09 | | 53.86 | 48.25 |
| Amag_GK30097_PI468340_UGA_B | 49.66 | 49.64 | 49.07 | 48.57 | 48.50 | 48.62 | 48.38 | 48.42 | 53.80 | 53.86 | | 55.47 |
| Ahoeh_VP9094_PI688954_Embrapa_B | 51.03 | 51.10 | 50.95 | 50.49 | 50.42 | 50.60 | 50.05 | 50.40 | 48.30 | 48.25 | 55.47 | |
| Amag_GK30097_PI468340_Embrapa2_B | 50.09 | 50.08 | 49.42 | 48.87 | 49.04 | 49.01 | 48.69 | 48.51 | 54.25 | 54.30 | 71.20 | 54.83 |
| Avali_VP13514_Embrapa_B | 49.08 | 49.04 | 48.77 | 48.31 | 48.17 | 48.21 | 48.08 | 47.98 | 56.16 | 56.05 | 65.80 | 55.49 |
| Amag_VS13751_Embrapa_B | 50.09 | 49.94 | 49.47 | 49.11 | 48.94 | 49.05 | 48.82 | 48.70 | 54.32 | 54.47 | 67.96 | 55.65 |
| Akemp_V14673_Embrapa_A | 52.73 | 52.65 | 51.79 | 52.01 | 52.06 | 52.02 | 51.86 | 52.26 | 49.75 | 49.59 | 53.41 | 61.04 |
| Ahoeh_V14547_Embrapa_B | 52.20 | 52.22 | 51.97 | 51.57 | 51.51 | 51.49 | 51.24 | 51.38 | 49.42 | 49.45 | 56.35 | 70.58 |
| Ahoeh_VP9140_PI666086_Embrapa_B | 51.91 | 52.07 | 51.89 | 51.42 | 51.26 | 51.31 | 50.84 | 51.04 | 49.11 | 49.29 | 56.27 | 70.34 |
| Amag_Se3777_IBONE_B | 50.01 | 50.01 | 49.38 | 49.04 | 48.99 | 49.08 | 48.66 | 48.71 | 54.48 | 54.53 | 68.94 | 55.84 |
| Avali_GK30011_PI468154_UGA_B | 49.35 | 49.39 | 49.17 | 48.75 | 48.56 | 48.73 | 48.44 | 48.34 | 56.73 | 56.76 | 66.58 | 55.76 |
| Ahoeh_VM13985_Embrapa_B | 51.80 | 51.84 | 51.70 | 51.19 | 51.20 | 51.24 | 50.88 | 50.84 | 49.22 | 49.27 | 56.03 | 70.89 |
| Amag_VS13761_Embrapa_B | 50.25 | 50.23 | 49.59 | 49.22 | 49.01 | 49.18 | 48.96 | 48.74 | 54.51 | 54.64 | 68.49 | 55.65 |
| Akemp_V14886_Embrapa_A | 54.19 | 54.05 | 53.04 | 53.62 | 53.41 | 53.32 | 53.52 | 53.46 | 50.82 | 50.91 | 53.55 | 61.67 |
Any help would be very appreciated,
Francisco