My code goes as follows:
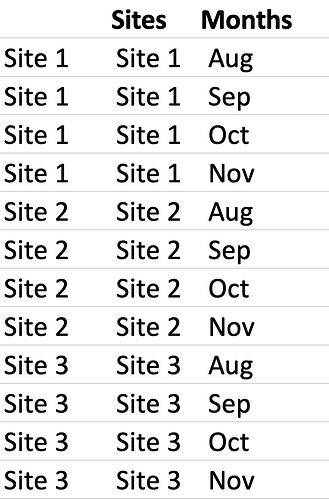
map <- structure(list(Sites = c("Site 1", "Site 1", "Site 1", "Site 1",
"Site 2", "Site 2", "Site 2", "Site 2", "Site 3", "Site 3", "Site 3",
"Site 3"), Months = c("Aug", "Sep", "Oct", "Nov", "Aug", "Sep",
"Oct", "Nov", "Aug", "Sep", "Oct", "Nov")),
class = "data.frame", row.names = c(NA, -12L))
# transform dataframe into phyloseq class object.
map <- sample_data(map)
otu.table <- read.csv(file = "mouse.csv", header = TRUE, sep = ",")
dput(otu.table)
structure(list(`Site 1` = c(1, 5, 1, 0, 0, 0, 0, 0, 3, 0, 0,
0, 0, 0, 0, 0, 0, 1, 2, 4, 2, 3, 2, 0, 0, 0, 0, 1, 1, 0, 0, 0,
0, 2, 0, 0, 5, 0), `Site 1` = c(0, 4, 0, 1, 0, 0, 0, 0, 8, 0,
0, 0, 0, 0, 0, 0, 2, 0, 2, 2, 0, 1, 0, 1, 3, 0, 0, 1, 2, 0, 0,
0, 1, 3, 2, 0, 1, 0), `Site 1` = c(5, 1, 0, 0, 0, 0, 0, 2, 11,
5, 0, 1, 2, 0, 0, 0, 0, 0, 2, 4, 1, 0, 0, 1, 0, 0, 0, 0, 0, 0,
0, 1, 0, 0, 0, 0, 0, 0), `Site 1` = c(9, 0, 0, 0, 1, 0, 0, 4,
3, 0, 0, 0, 0, 0, 0, 0, 1, 5, 2, 7, 0, 0, 0, 3, 0, 0, 0, 1, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0), `Site 2` = c(0, 0, 0, 0, 0, 0, 0,
2, 3, 0, 1, 3, 0, 0, 0, 0, 0, 1, 0, 9, 6, 0, 1, 0, 0, 0, 0, 0,
2, 0, 0, 0, 0, 0, 0, 0, 0, 0), `Site 2` = c(1, 0, 0, 0, 0, 4,
0, 4, 2, 0, 1, 3, 0, 3, 0, 1, 0, 1, 0, 9, 0, 0, 3, 0, 1, 0, 0,
0, 0, 2, 0, 0, 0, 0, 1, 0, 0, 1), `Site 2` = c(13, 0, 0, 0, 0,
0, 0, 0, 1, 1, 2, 1, 0, 0, 0, 0, 0, 1, 0, 8, 1, 0, 2, 0, 0, 0,
0, 0, 0, 0, 0, 1, 0, 0, 1, 1, 0, 0), `Site 2` = c(12, 2, 0, 0,
1, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 1, 9, 0, 0, 2, 0, 3,
0, 0, 0, 0, 0, 0, 1, 0, 1, 0, 0, 0, 5), `Site 3` = c(6, 0, 0,
0, 0, 0, 0, 1, 4, 1, 0, 0, 0, 0, 0, 0, 1, 0, 1, 9, 2, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 5, 1, 0, 0, 0, 0, 2), `Site 3` = c(5, 1,
0, 0, 0, 0, 0, 3, 5, 0, 0, 2, 0, 0, 0, 0, 0, 0, 3, 7, 1, 0, 0,
0, 0, 2, 0, 0, 3, 1, 0, 5, 0, 0, 0, 0, 0, 0), `Site 3` = c(14,
0, 0, 0, 2, 0, 0, 0, 1, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 3, 0, 0,
0, 1, 0, 0, 0, 0, 3, 1, 0, 4, 2, 0, 1, 1, 0, 2), `Site 3` = c(22,
0, 0, 0, 0, 0, 0, 4, 2, 0, 0, 0, 0, 0, 0, 0, 1, 1, 0, 1, 0, 0,
1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0)), row.names = c("otu001",
"otu002", "otu003", "otu004", "otu005", "otu006", "otu007", "otu008",
"otu009", "otu010", "otu011", "otu012", "otu013", "otu014", "otu015",
"otu016", "otu017", "otu018", "otu019", "otu020", "otu021", "otu022",
"otu023", "otu024", "otu025", "otu026", "otu027", "otu028", "otu029",
"otu030", "otu031", "otu032", "otu033", "otu034", "otu035", "otu036",
"otu037", ""), class = "data.frame")
rownames(otu.table)<- otu.table$otus
otu.table <- otu.table[, colnames(otu.table) != "otus"]
colnames(otu.table) <- c("Site 1", "Site 1", "Site 1", "Site 1",
"Site 2", "Site 2", "Site 2", "Site 2", "Site 3", "Site 3", "Site 3",
"Site 3")
otu.table[is.na(otu.table)] <- 0
otu_mat <- as.matrix(otu.table)
# transform dataframe into phyloseq class object.
otu_mat <- otu_table(otu_mat, taxa_are_rows = T)
Phyloseq_object <- merge_phyloseq(otu_mat, map)
My mapfile looks like this
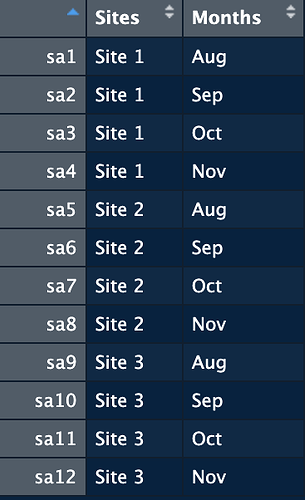
The row names are not correct here.
When I use
merge_phyloseq(otu_mat, map)
I get this error:
Error in validObject(.Object) : invalid class “phyloseq” object:
Component sample names do not match.
Try sample_names()
And I cannot proceed further.
Duplicating rownames are possible. You can check the code here:
I tried it on mouse.dpw.metadata of this link(https://zenodo.org/record/800651#.ZBbaCuxBxJU).
It works but not on my metadata file. I cannot figure it out.
I hope I was able to explain my query clearly.
regards
Hira