Hi all ! I am trying to create a geom extension. I have the following issue :
I expected setup_data to add the required_aes in the data and then pass it to draw_group
but the setup_data method fails as it enters draw_group method and throws the above mentioned error.
Please offer some suggestions !
the data after setup_params looks like this
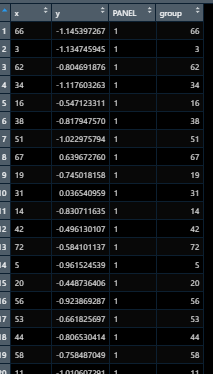
The data just before setup_data finishes
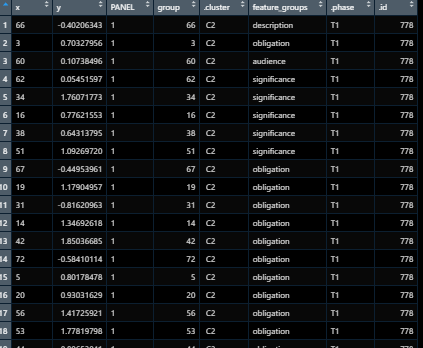
What the data should like : 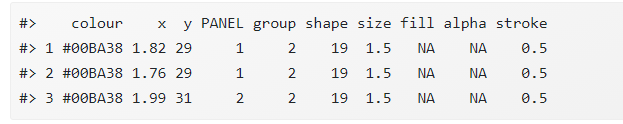
GeomRbar <- ggplot2::ggproto("GeomRbar", ggplot2::Geom,
required_aes = c("x", "y"),
default_aes = ggplot2::aes(
fill = "grey60",
size = 1.25,
colour_inner_circle = "grey90",
standard_error = TRUE
),
setup_data = function(data, params) {
browser()
data <- data %>%
dplyr::mutate(.cluster = params$cluster_assignment) %>%
dplyr::mutate(feature_groups = params$group_names)
if (!is.null(params$cluster_phase)) {
data <- data %>% dplyr::mutate(.phase = params$cluster_phase)
} else {
data <- data %>% dplyr::mutate(.phase := NA_character_)
}
if (!is.null(params$unique_id)) {
data <- data %>% dplyr::mutate(.id = params$unique_id)
} else {
data <- data %>% mutate(.id := NA_character_)
}
vars_dummy <- setdiff(c(".phase", ".id", ".cluster", "feature_groups"), names(data))
if (length(vars_dummy) > 0) {
rlang::abort("Data doesn't have required structure to build the plot")
}
#### Calculate cluster average for each feature
if (!is.null(params$cluster_idx)) {
rlang::inform("Calculating cluster average for each feature")
data <- data %>% dplyr::filter(.cluster == levels(.cluster)[params$cluster_idx]) # now contains observations from one cluster(cluster of interest) e.g C2 has 23713 observations
}
if (!is.null(params$cluster_idx)) {
data <- data %>%
dplyr::mutate(.cluster = forcats::fct_drop(as.factor(.cluster))) %>% # fct_drop - drops unused levels
dplyr::mutate(.cluster = factor(.cluster, labels = paste(params$cluster_abbrev, levels(.cluster)))) # add cluster_abbrev to .cluster e.g : PT C2
} else {
data <- data
}
browser()
},
# draw_group ----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
draw_group = function(data, params, cluster_idx = NULL,
cluster_assignment = NULL,
phase_present = FALSE,
cluster_phase = NULL,
cluster_name = NULL,
cluster_abbrev = NULL,
colour_clusters = NULL,
scale_rng = c(-1, 1) * 1.5,
data_dict = NULL,
delta_threshold = 0.25,
group_names = NULL,
show_group_names = FALSE,
unique_id = NULL,
na.rm = FALSE) {
data_check <- data
browser()