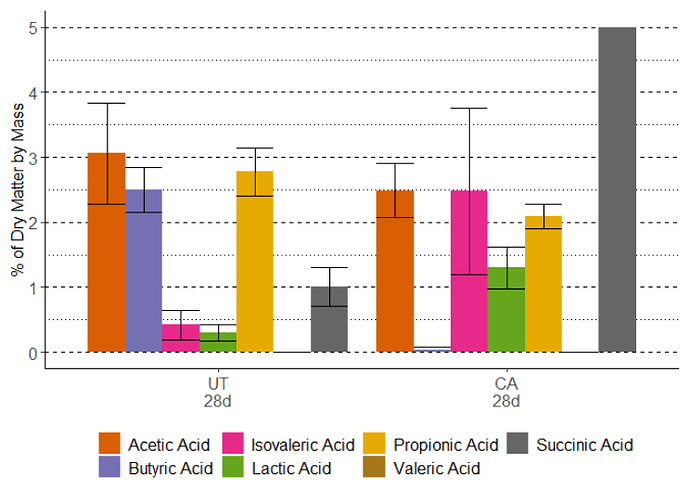
So I'm trying to make a histogram with ggplot where I am trying to zoom in the axis on the a graph, but I still want all my bars to be present. So I have the dataframe below:
> dput(organic_acids_28day_merged_numeric)
structure(list(Sample = c("Ana 28day", "Ana 28day", "Ana 28day",
"Ana 28day", "Ana 28day", "Ana 28day", "Ana 28day", "CA 28day",
"CA 28day", "CA 28day", "CA 28day", "CA 28day", "CA 28day", "CA 28day"
), Acid = c("Acetic Acid", "Butyric Acid", "Isovaleric Acid",
"Lactic Acid", "Propionic Acid", "Succinic Acid", "Valeric Acid",
"Acetic Acid", "Butyric Acid", "Isovaleric Acid", "Lactic Acid",
"Propionic Acid", "Succinic Acid", "Valeric Acid"), Percent = c(3.06,
2.5, 0.42, 0.3, 2.78, 1.01, 0, 2.49, 0.03, 2.48, 1.3, 2.09, 29.68,
0), stdev = c(0.78, 0.34, 0.23, 0.13, 0.37, 0.3, 0, 0.41, 0.05,
1.28, 0.32, 0.19, 6.81, 0)), class = "data.frame", row.names = c(NA,
-14L))
And when I don't try to fix the axes I get all the data points showing up fine. But when I zoom into the axis, as with the code below (adjusting the y-axis to be betwen 0 and 5) it completely removes the bar that goes up to about 30%. And I get the error after it:
> ggplot(organic_acids_28day_merged_numeric,
+ aes(x=factor(Sample,level=c("Ana 28day","CA 28day")),
+ y=Percent,fill=factor(Acid,level=c("Acetic Acid",
+ 'Butyric Acid',
+ 'Isovaleric Acid',
+ 'Lactic Acid',
+ 'Propionic Acid',
+ 'Valeric Acid',
+ 'Succinic Acid'))))+
+ geom_col(position=position_dodge())+
+ labs(x="None",y="% of Dry Matter by Mass")+
+ scale_x_discrete(labels=c("UT\n28d","CA\n28d"))+
+ scale_y_continuous(limits=c(0,5))+
+ geom_errorbar(aes(ymin=Percent-stdev,
+ ymax=Percent+stdev),
+ position=position_dodge())+
+ scale_fill_manual(values=c("#D95F02","#7570B3","#E7298A","#66A61E","#E6AB02",
+ "#A6761D","#666666"))+
+ # scale_fill_brewer(palette="Dark2")+
+ theme_classic()+
+ theme(axis.title.x=element_blank(),legend.title=element_blank(),
+ legend.text =element_text(size=12),axis.title.y=element_text(size=12),
+ legend.position="bottom",axis.text.x=element_text(size=12),
+ axis.text.y=element_text(size=12),
+ panel.grid.major.y=element_line(color=1,size=0.5,linetype=2),
+ panel.grid.minor.y=element_line(color="black",size=0.25,linetype=3))
Warning message:
Removed 1 rows containing missing values (`geom_col()`).
And I know it's removed because the value is outside the y-axis limit. But I wanted to know if there is a way to fix this so that it still shows that last bar being present, even if it would be cut off, if that makes sense.