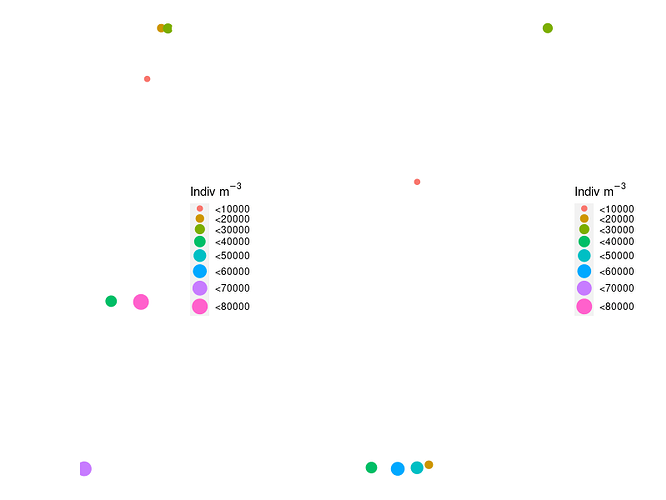
Hello,
I have two plots with different data ranges. I want them to have the same legend with the same colors associated with their categorical variables. I am trying to force my ggplot2 legend to show categorical values that are not associated with any point. There used to be an argument called drop = FALSE in scale_color_manual but that no longer works. Does anybody know the argument to use to force ggplot2 to show unused variables in the legend?
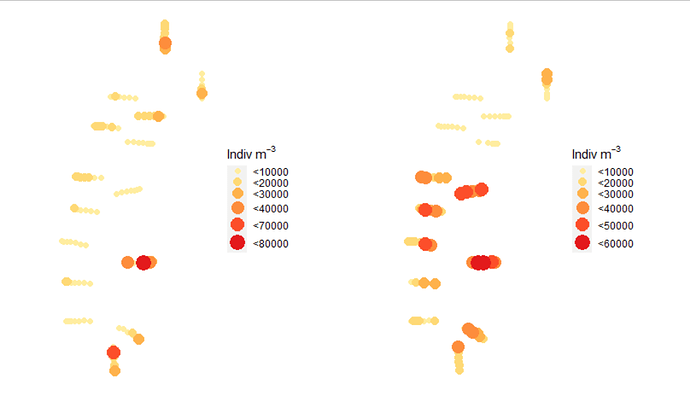
Here is my plot:
See how the legend scales are not the same?
Here is a subset of the data that I used (does not show all points):
data <- structure(list(TRANSECT = c("EAS12", "EAS12", "EMP12", "EMP17",
"GH12", "GH17", "GH17", "GH23", "GH38", "GH38", "GH5", "GTB30",
"MIC30"), Group = c("Cladoceran", "Cladoceran", "Rotifera", "Rotifera",
"Rotifera", "Rotifera", "Mollusks", "Rotifera", "Rotifera", "Rotifera",
"Rotifera", "Rotifera", "Rotifera"), DEPTH_CLASS = c("UV", "No UV",
"UV", "UV", "No UV", "UV", "No UV", "No UV", "UV", "No UV", "No UV",
"No UV", "UV"), Density = c(2.292795622, 72.11726105, 22025.72841,
11004.65472, 46620.94058, 74660.99802, NA, 50434.93142, 31305.67666,
33942.05725, 12871.3228, 20507.28229, 66078.24687), geometry = structure(list(
structure(c(-86.4035333, 44.49025), class = c("XY", "POINT",
"sfg")), structure(c(-86.4035333, 44.49025), class = c("XY",
"POINT", "sfg")), structure(c(-86.2197667, 44.8151167), class = c("XY",
"POINT", "sfg")), structure(c(-86.2789333, 44.8171167), class = c("XY",
"POINT", "sfg")), structure(c(-86.40435, 43.059567), class = c("XY",
"POINT", "sfg")), structure(c(-86.458617, 43.055617), class = c("XY",
"POINT", "sfg")), structure(c(-86.458617, 43.055617), class = c("XY",
"POINT", "sfg")), structure(c(-86.539017, 43.054067), class = c("XY",
"POINT", "sfg")), structure(c(-86.722433, 43.060333), class = c("XY",
"POINT", "sfg")), structure(c(-86.722433, 43.060333), class = c("XY",
"POINT", "sfg")), structure(c(-86.322017, 43.074533), class = c("XY",
"POINT", "sfg")), structure(c(-85.4930167, 45.2598), class = c("XY",
"POINT", "sfg")), structure(c(-86.96095, 41.981917), class = c("XY",
"POINT", "sfg"))), class = c("sfc_POINT", "sfc"), precision = 0, bbox = structure(c(xmin = -86.96095,
ymin = 41.981917, xmax = -85.4930167, ymax = 45.2598), class = "bbox"), crs = structure(list(
input = "EPSG:4326", wkt = "GEOGCRS[\"WGS 84\",\n ENSEMBLE[\"World Geodetic System 1984 ensemble\",\n MEMBER[\"World Geodetic System 1984 (Transit)\"],\n MEMBER[\"World Geodetic System 1984 (G730)\"],\n MEMBER[\"World Geodetic System 1984 (G873)\"],\n MEMBER[\"World Geodetic System 1984 (G1150)\"],\n MEMBER[\"World Geodetic System 1984 (G1674)\"],\n MEMBER[\"World Geodetic System 1984 (G1762)\"],\n MEMBER[\"World Geodetic System 1984 (G2139)\"],\n ELLIPSOID[\"WGS 84\",6378137,298.257223563,\n LENGTHUNIT[\"metre\",1]],\n ENSEMBLEACCURACY[2.0]],\n PRIMEM[\"Greenwich\",0,\n ANGLEUNIT[\"degree\",0.0174532925199433]],\n CS[ellipsoidal,2],\n AXIS[\"geodetic latitude (Lat)\",north,\n ORDER[1],\n ANGLEUNIT[\"degree\",0.0174532925199433]],\n AXIS[\"geodetic longitude (Lon)\",east,\n ORDER[2],\n ANGLEUNIT[\"degree\",0.0174532925199433]],\n USAGE[\n SCOPE[\"Horizontal component of 3D system.\"],\n AREA[\"World.\"],\n BBOX[-90,-180,90,180]],\n ID[\"EPSG\",4326]]"), class = "crs"), n_empty = 0L),
Dens_CAT = c("<10000", "<10000", "<30000", "<20000", "<50000",
"<80000", NA, "<60000", "<40000", "<40000", "<20000", "<30000",
"<70000")), class = c("sf", "grouped_df", "tbl_df", "tbl",
"data.frame"), row.names = c(NA, -13L), groups = structure(list(
DEPTH_CLASS = c("No UV", "UV"), .rows = structure(list(c(2L,
5L, 7L, 8L, 10L, 11L, 12L), c(1L, 3L, 4L, 6L, 9L, 13L)), ptype = integer(0), class = c("vctrs_list_of",
"vctrs_vctr", "list"))), class = c("tbl_df", "tbl", "data.frame"
), row.names = c(NA, -2L), .drop = TRUE), sf_column = "geometry", agr = structure(c(TRANSECT = NA_integer_,
Group = NA_integer_, DEPTH_CLASS = NA_integer_, Density = NA_integer_,
Dens_CAT = NA_integer_), class = "factor", levels = c("constant",
"aggregate", "identity"))
Here is the code that I'm using to generate the plots
themes <- theme(axis.text.x = element_blank(), # remove x-axis text
axis.text.y = element_blank(), # remove y-axis text
axis.ticks = element_blank(), # remove axis ticks
axis.title.x = element_text(size=18), # remove x-axis labels
axis.title.y = element_text(size=18), # remove y-axis labels
panel.background = element_blank(),
panel.grid.major = element_blank(), #remove major-grid labels
panel.grid.minor = element_blank(), #remove minor-grid labels
plot.background = element_blank(),
legend.key.size = unit(0.25, 'cm'))
p1 <- data %>%
filter(DEPTH_CLASS == "UV") %>%
drop_na(Dens_CAT) %>%
ggplot() +
geom_sf(aes(size = Dens_CAT, color=Dens_CAT)) +
scale_color_discrete(type = cols,
drop = FALSE) + # this argument does not work
themes +
guides(color='legend', size='legend') +
labs(color = bquote('Indiv'~m^-3),
size = bquote('Indiv'~m^-3))
p2 <- data %>%
filter(DEPTH_CLASS == "No UV") %>%
drop_na(Dens_CAT) %>%
ggplot() +
geom_sf(aes(size = Dens_CAT, color=Dens_CAT)) +
scale_color_discrete(type = cols,
drop = FALSE) + #this argument does not work
themes +
guides(color='legend', size='legend') +
labs(color = bquote('Indiv'~m^-3),
size = bquote('Indiv'~m^-3))
grid.arrange(p1,p2, nrow=1)
Any help would be greatly appreciated. Thank you so much!