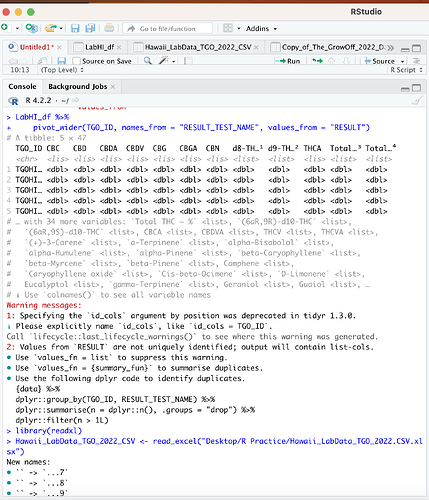
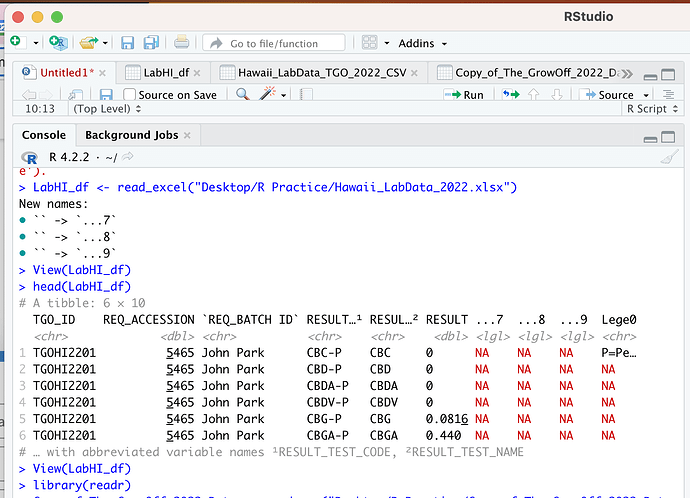
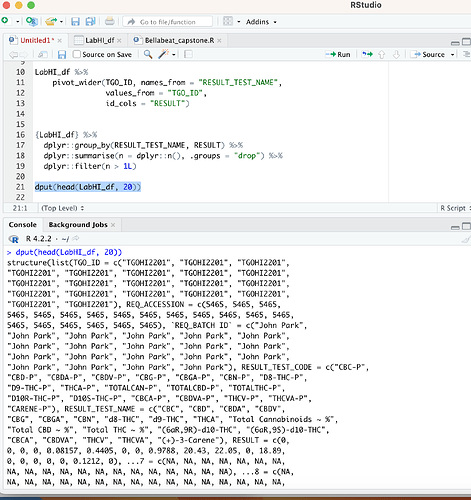
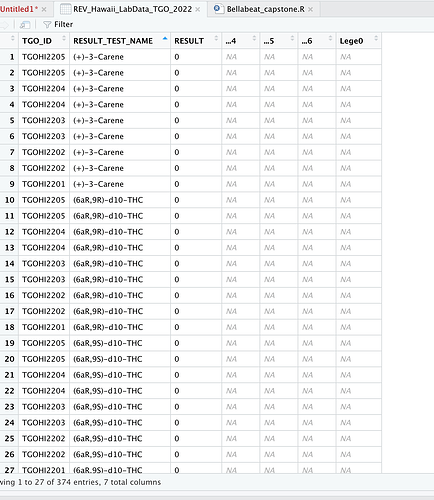
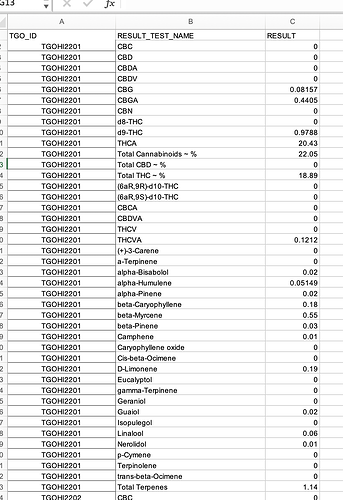
The problem is RESULT_TEST_NAME has repeated values for some TGO_ID if I select another column with unique values like RESULT_TEST_CODE it works just fine. Otherwise, you would need to decide what to do with repeated values per RESULT_TEST_NAME/TGO_ID combination.
library(dplyr)
library(tidyr)
# Sample data (replace this with your own data)
Hawaii_LabData_2022 <- structure(list(TGO_ID = c("TGOHI2201", "TGOHI2201", "TGOHI2201",
"TGOHI2201", "TGOHI2201", "TGOHI2201", "TGOHI2201", "TGOHI2201",
"TGOHI2201", "TGOHI2201", "TGOHI2201", "TGOHI2201", "TGOHI2201",
"TGOHI2201", "TGOHI2201", "TGOHI2201", "TGOHI2201", "TGOHI2201",
"TGOHI2201", "TGOHI2201", "TGOHI2201", "TGOHI2201", "TGOHI2201",
"TGOHI2201", "TGOHI2201", "TGOHI2201", "TGOHI2201", "TGOHI2201",
"TGOHI2201", "TGOHI2201", "TGOHI2201", "TGOHI2201", "TGOHI2201",
"TGOHI2201", "TGOHI2201", "TGOHI2201", "TGOHI2201", "TGOHI2201",
"TGOHI2201", "TGOHI2201", "TGOHI2201", "TGOHI2201", "TGOHI2202",
"TGOHI2202", "TGOHI2202", "TGOHI2202", "TGOHI2202", "TGOHI2202",
"TGOHI2202", "TGOHI2202", "TGOHI2202", "TGOHI2202", "TGOHI2202",
"TGOHI2202", "TGOHI2202", "TGOHI2202", "TGOHI2202", "TGOHI2202",
"TGOHI2202", "TGOHI2202", "TGOHI2202", "TGOHI2202", "TGOHI2202",
"TGOHI2202", "TGOHI2202", "TGOHI2202", "TGOHI2202", "TGOHI2202",
"TGOHI2202", "TGOHI2202", "TGOHI2202", "TGOHI2202", "TGOHI2202",
"TGOHI2202", "TGOHI2202", "TGOHI2202", "TGOHI2202", "TGOHI2202",
"TGOHI2202", "TGOHI2202", "TGOHI2202", "TGOHI2202", "TGOHI2202",
"TGOHI2202", "TGOHI2202", "TGOHI2202", "TGOHI2202", "TGOHI2202",
"TGOHI2202", "TGOHI2202", "TGOHI2202", "TGOHI2202", "TGOHI2202",
"TGOHI2202", "TGOHI2202", "TGOHI2202", "TGOHI2202", "TGOHI2202",
"TGOHI2202", "TGOHI2202"), REQ_ACCESSION = c(5465, 5465, 5465,
5465, 5465, 5465, 5465, 5465, 5465, 5465, 5465, 5465, 5465, 5465,
5465, 5465, 5465, 5465, 5465, 5465, 5465, 5465, 5465, 5465, 5465,
5465, 5465, 5465, 5465, 5465, 5465, 5465, 5465, 5465, 5465, 5465,
5465, 5465, 5465, 5465, 5465, 5465, 5484, 5484, 5484, 5484, 5484,
5484, 5484, 5484, 5484, 5484, 5484, 5484, 5484, 5484, 5484, 5484,
5484, 5484, 5484, 5484, 5484, 5484, 5484, 5484, 5484, 5484, 5484,
5484, 5484, 5484, 5484, 5484, 5484, 5484, 5484, 5484, 5484, 5484,
5484, 5484, 5484, 5484, 5484, 5484, 5484, 5484, 5484, 5484, 5484,
5484, 5484, 5484, 5484, 5484, 5484, 5484, 5484, 5484), REQ_BATCH_ID = c("John Park",
"John Park", "John Park", "John Park", "John Park", "John Park",
"John Park", "John Park", "John Park", "John Park", "John Park",
"John Park", "John Park", "John Park", "John Park", "John Park",
"John Park", "John Park", "John Park", "John Park", "John Park",
"John Park", "John Park", "John Park", "John Park", "John Park",
"John Park", "John Park", "John Park", "John Park", "John Park",
"John Park", "John Park", "John Park", "John Park", "John Park",
"John Park", "John Park", "John Park", "John Park", "John Park",
"John Park", "Jan Flores", "Jan Flores", "Jan Flores", "Jan Flores",
"Jan Flores", "Jan Flores", "Jan Flores", "Jan Flores", "Jan Flores",
"Jan Flores", "Jan Flores", "Jan Flores", "Jan Flores", "Jan Flores",
"Jan Flores", "Jan Flores", "Jan Flores", "Jan Flores", "Jan Flores",
"Jan Flores", "Jan Flores", "Jan Flores", "Jan Flores", "Jan Flores",
"Jan Flores", "Jan Flores", "Jan Flores", "Jan Flores", "Jan Flores",
"Jan Flores", "Jan Flores", "Jan Flores", "Jan Flores", "Jan Flores",
"Jan Flores", "Jan Flores", "Jan Flores", "Jan Flores", "Jan Flores",
"Jan Flores", "Jan Flores", "Jan Flores", "Jan Flores", "Jan Flores",
"Jan Flores", "Jan Flores", "Jan Flores", "Jan Flores", "Jan Flores",
"Jan Flores", "Jan Flores", "Jan Flores", "Jan Flores", "Jan Flores",
"Jan Flores", "Jan Flores", "Jan Flores", "Jan Flores"), RESULT_TEST_CODE = c("CBC-P",
"CBD-P", "CBDA-P", "CBDV-P", "CBG-P", "CBGA-P", "CBN-P", "D8-THC-P",
"D9-THC-P", "THCA-P", "TOTALCAN-P", "TOTALCBD-P", "TOTALTHC-P",
"D10R-THC-P", "D10S-THC-P", "CBCA-P", "CBDVA-P", "THCV-P", "THCVA-P",
"CARENE-P", "A_TERPINENE_P", "A_BISABOLOL-P", "A_HUMULENE-P",
"A_PINENE-P", "B_CARYOPHYLLENE-P", "B_MYRCENE-P", "B_PINENE-P",
"CAMPHENE-P", "CARYOPHYLLENE_OXIDE-P", "C_OCIMENE-P", "D_LIMONENE-P",
"EUCALYPTOL-P", "G_TERPINENE-P", "GERANIOL-P", "GUAIOL-P", "ISOPULEGOL-P",
"LINALOOL-P", "NEROLIDOL-P", "P_CYMENE-P", "TERPINOLENE-P", "T_OCIMENE-P",
"Total Terpenes", "CBC-P", "CBC-MGG", "CBD-MGG", "CBD-P", "CBDA-P",
"CBDA-MGG", "CBDV-MGG", "CBDV-P", "CBG-P", "CBG-MGG", "CBGA-MGG",
"CBGA-P", "CBN-P", "CBN-MGG", "D8-THC-MGG", "D8-THC-P", "D9-THC-P",
"D9-THC-MGG", "THCA-MGG", "THCA-P", "TOTALCAN-P", "TOTALCAN-MGG",
"TOTALCBD-P", "TOTALCBD-MGG", "TOTALTHC-P", "TOTALTHC-MGG", "D10R-THC-P",
"D10R-THC-MGG", "D10S-THC-MGG", "D10S-THC-P", "CBCA-MGG", "CBCA-P",
"CBDVA-P", "CBDVA-MGG", "THCV-P", "THCV-MGG", "THCVA-MGG", "THCVA-P",
"CARENE-P", "CARENE-MGG", "A_TERPINENE_P", "A_TERPINENE-MGG",
"A_BISABOLOL-MGG", "A_BISABOLOL-P", "A_HUMULENE-P", "A_HUMULENE-MGG",
"A_PINENE-P", "A_PINENE-MGG", "B_CARYOPHYLLENE-MGG", "B_CARYOPHYLLENE-P",
"B_MYRCENE-P", "B_MYRCENE-MGG", "B_PINENE-P", "B_PINENE-MGG",
"CAMPHENE-MGG", "CAMPHENE-P", "CARYOPHYLLENE_OXIDE-MGG", "CARYOPHYLLENE_OXIDE-P"
), RESULT_TEST_NAME = c("CBC", "CBD", "CBDA", "CBDV", "CBG",
"CBGA", "CBN", "d8-THC", "d9-THC", "THCA", "Total Cannabinoids ~ %",
"Total CBD ~ %", "Total THC ~ %", "(6aR,9R)-d10-THC", "(6aR,9S)-d10-THC",
"CBCA", "CBDVA", "THCV", "THCVA", "(+)-3-Carene", "a-Terpinene",
"alpha-Bisabolol", "alpha-Humulene", "alpha-Pinene", "beta-Caryophyllene",
"beta-Myrcene", "beta-Pinene", "Camphene", "Caryophyllene oxide",
"Cis-beta-Ocimene", "D-Limonene", "Eucalyptol", "gamma-Terpinene",
"Geraniol", "Guaiol", "Isopulegol", "Linalool", "Nerolidol",
"p-Cymene", "Terpinolene", "trans-beta-Ocimene", "Total Terpenes",
"CBC", "CBC", "CBD", "CBD", "CBDA", "CBDA", "CBDV", "CBDV", "CBG",
"CBG", "CBGA", "CBGA", "CBN", "CBN", "d8-THC", "d8-THC", "d9-THC",
"d9-THC", "THCA", "THCA", "Total Cannabinoids ~ %", "Total Cannabinoids ~ mg/g",
"Total CBD ~ %", "Total CBD ~ mg/g", "Total THC ~ %", "Total THC ~ mg/g",
"(6aR,9R)-d10-THC", "(6aR,9R)-d10-THC", "(6aR,9S)-d10-THC", "(6aR,9S)-d10-THC",
"CBCA", "CBCA", "CBDVA", "CBDVA", "THCV", "THCV", "THCVA", "THCVA",
"(+)-3-Carene", "(+)-3-Carene", "a-Terpinene", "a-Terpinene",
"alpha-Bisabolol", "alpha-Bisabolol", "alpha-Humulene", "alpha-Humulene",
"alpha-Pinene", "alpha-Pinene", "beta-Caryophyllene", "beta-Caryophyllene",
"beta-Myrcene", "beta-Myrcene", "beta-Pinene", "beta-Pinene",
"Camphene", "Camphene", "Caryophyllene oxide", "Caryophyllene oxide"
), RESULT = c(0, 0, 0, 0, 0.08157, 0.4405, 0, 0, 0.9788, 20.43,
22.05, 0, 18.89, 0, 0, 0, 0, 0, 0.1212, 0, 0, 0.02, 0.05149,
0.02, 0.18, 0.55, 0.03, 0.01, 0, 0, 0.19, 0, 0, 0, 0.02, 0, 0.06,
0.01, 0, 0, 0, 1.14, 0, 0, 0, 0, 0, 0, 0, 0, 0.1266, 1.266, 9.365,
0.9365, 0, 0, 0, 0, 0.4853, 4.853, 145.9, 14.59, 16.14, 161.4,
0, 0, 13.28, 132.8, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0.27, 0.03, 0.04535, 0.45, 0.03, 0.25, 1.65, 0.17, 0.2,
2.01, 0.04, 0.39, 0.07, 0.01, 0, 0), ...7 = c(NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA
), ...8 = c(NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA), ...9 = c(NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA), Lege0 = c("P=Percent",
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA)), row.names = c(NA, -100L), class = c("tbl_df", "tbl",
"data.frame"))
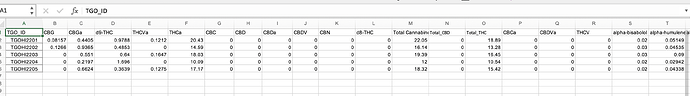
Hawaii_LabData_2022 %>%
pivot_wider(id_cols = TGO_ID, names_from = RESULT_TEST_CODE, values_from = RESULT)
#> # A tibble: 2 × 72
#> TGO_ID `CBC-P` `CBD-P` CBDA-…¹ CBDV-…² `CBG-P` CBGA-…³ `CBN-P` D8-TH…⁴ D9-TH…⁵
#> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 TGOHI… 0 0 0 0 0.0816 0.440 0 0 0.979
#> 2 TGOHI… 0 0 0 0 0.127 0.936 0 0 0.485
#> # … with 62 more variables: `THCA-P` <dbl>, `TOTALCAN-P` <dbl>,
#> # `TOTALCBD-P` <dbl>, `TOTALTHC-P` <dbl>, `D10R-THC-P` <dbl>,
#> # `D10S-THC-P` <dbl>, `CBCA-P` <dbl>, `CBDVA-P` <dbl>, `THCV-P` <dbl>,
#> # `THCVA-P` <dbl>, `CARENE-P` <dbl>, A_TERPINENE_P <dbl>,
#> # `A_BISABOLOL-P` <dbl>, `A_HUMULENE-P` <dbl>, `A_PINENE-P` <dbl>,
#> # `B_CARYOPHYLLENE-P` <dbl>, `B_MYRCENE-P` <dbl>, `B_PINENE-P` <dbl>,
#> # `CAMPHENE-P` <dbl>, `CARYOPHYLLENE_OXIDE-P` <dbl>, `C_OCIMENE-P` <dbl>, …
Created on 2023-02-23 with reprex v2.0.2