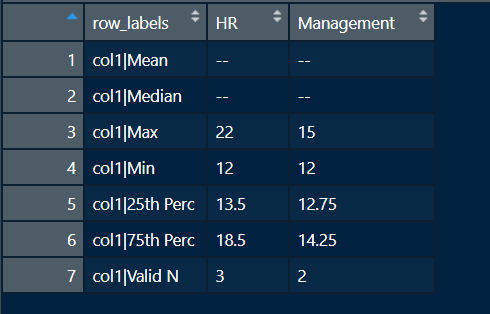
I am looking to hide values from the output table if the frequency of data in the respective variable is less than 4 .
lets say if the number of records in column hp, mpg, qsec is less than 4 than the mean or median should be masked with "--"
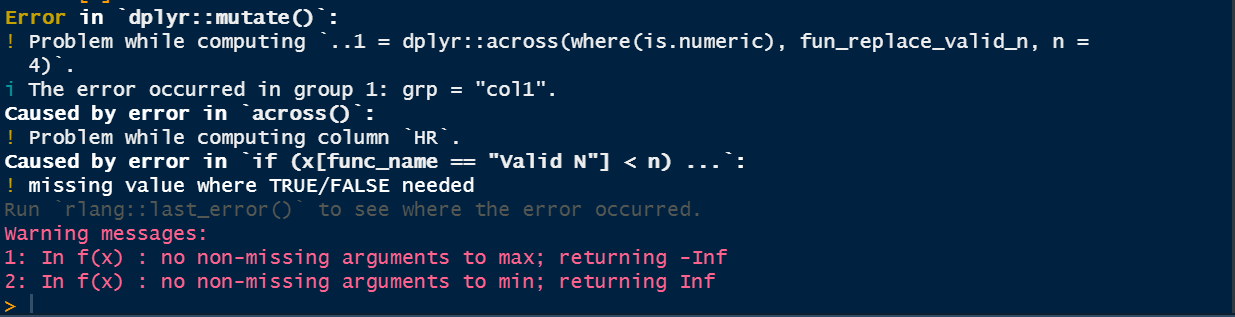
i am trying like below but not working showing some error due to NA in database
library(expss)
data <- data.frame(
gender = c(1, 2, 1),
sector = c(3, 3, 1),
col1 = c(12, 15, 22),
col2 = c(33, NA, 41),
col3 = c(1, 1, 0),
col4 = c(NA,NA,NA),
col5 = c(1, 2, 1)
)
data$col3 <- factor(data$col3, levels = 1, labels = "Management")
data$col4 <- factor(data$col4, levels = 1, labels = "HR")
lst <- list(data$col4,data$col3)
fun1 <- function(dataset,var_list,banner1){
perc_25 <- function(x, ...){unname(quantile(x, .25, na.rm=TRUE))}
perc_75 <- function(x, ...){unname(quantile(x, .75, na.rm=TRUE))}
dataset<-dataset[var_list] %>% as.data.frame()
first_col_param <- head(var_list,1)
second_col_param <- tail(var_list,1)
var_lab(colnames(dataset)[ncol(dataset)]) <- ""
mr <- parse(text=paste0("mrset(",
first_col_param ," %to% ",second_col_param,")"))
fun_replace_valid_n <- function(x, n) {
dat <- dplyr::cur_data_all() %>% replace(is.na(.),0)
func_name <- dat$func_name
if(x[func_name == "Valid N"] < n) {
replace(x, func_name %in% c("Mean", "Median"), "--")
} else x
}
t1<- cross_fun(dataset,
eval(mr),
col_vars = banner1,
fun = combine_functions("Mean" = mean,
"Median" = median,
"Max"= max,
"Min"=min,
"25th Perc" = perc_25,
"75th Perc" = perc_75,
"Valid N" = valid_n
))
t1 <- as.data.frame(t1)
t1 <- t1 %>% tidyr::separate(row_labels, into = c('grp', 'func_name'), sep = "\\|")
t1 <- t1 %>% dplyr::group_by(grp)
t1 <- t1 %>% dplyr::mutate(dplyr::across(where(is.numeric), fun_replace_valid_n, n = 4)) %>%
dplyr::ungroup()
t1 <- t1 %>% tidyr::unite(row_labels, grp, func_name, sep = "|") %>%
as.etable
t1
}
debugonce(fun1)
t1 <- fun1(dataset=data,"col1",banner1=lst)
THe output should be look like