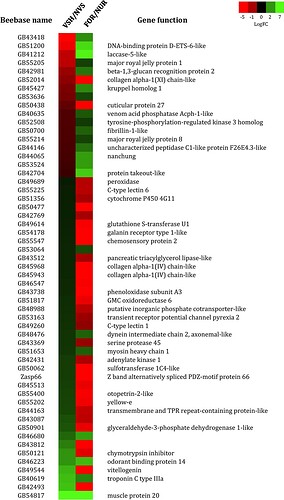
Hello everyone, I have doubts about how to create a heat map. There is a lot of information on the internet on how to create them but basically it is very simple information, with little data. In my case, I would like to make a heat map that has the associated genes, with their description of the genes, and that is compared between months. I mean something like this:
And I think I have the whole information I need for this kind of graphs.
Data <- data.frame (tibble::tribble(
~Beebase_name, ~Description, ~logFCJULVSAUG, ~logFCJULVSSEPT, ~logFCAUGVSSEPT,
"map04932", "Non-alcoholic fatty liver disease", 0.075670032, 0.003935337, 0.01408051,
"map00332", "Carbapenem biosynthesis", -0.014031521, 4.9e-05, -0.071734739,
"map00333", "Prodigiosin biosynthesis", 0.007131704, 0.020343039, 0.013211291,
"map04933", "AGE-RAGE signaling pathway in diabetic complications", 0.004932971, -0.051448915, -0.05638193,
"map00330", "Arginine and proline metabolism", 0.010045605, -0.003979007, -0.014024656,
"map04934", "Cushing syndrome", 0.010501308, -0.006209653, -0.016711005,
"map00331", "Clavulanic acid biosynthesis", -0.066802306, -0.079918417, -0.013116154,
"map00450", "Selenocompound metabolism", -0.002724964, -0.00292324, -0.000198,
"map04930", "Type II diabetes mellitus", 0.004425738, -0.001000992, -0.005426774,
"map04931", "Insulin resistance", -0.003641615, -0.000937, 0.002704848,
"map04810", "Regulation of actin cytoskeleton", 0.316469292, -0.126381822, -0.442851157,
"map00563", "Glycosylphosphatidylinositol (GPI)-anchor biosynthesis", 0.172221706, -0.074206148, -0.615193702,
"map04921", "Oxytocin signaling pathway", 0.356132843, -0.259060815, -0.246427898,
"map00564", "Glycerophospholipid metabolism", -0.012186907, -0.008827405, 0.003359457,
"map04922", "Glucagon signaling pathway", 0.000999, -0.005950848, -0.006949856,
"map00561", "Glycerolipid metabolism", 0.002776084, 0.005820994, 0.003044867,
"map00440", "Phosphonate and phosphinate metabolism", 0.103317267, 0.05697387, -0.526828638,
"map04923", "Regulation of lipolysis in adipocytes", 0.40709608, -0.119732514, -0.046343441,
"map00562", "Inositol phosphate metabolism", 0.00227146, -0.006225672, -0.008497176,
"map04924", "Renin secretion", 0.005229426, -0.313909259, -0.319138729,
"map00680", "Methane metabolism", -0.002860128, 0.017217665, 0.020077748,
"map04925", "Aldosterone synthesis and secretion", 0.113626918, -0.327363593, -0.440990554,
"map04926", "Relaxin signaling pathway", 0.003958041, -0.095366001, -0.099324086,
"map04927", "Cortisol synthesis and secretion", 0.125877766, -0.475589127, -0.601466936
)
)
Created on 2022-01-27 by the reprex package (v2.0.1)