Hi,
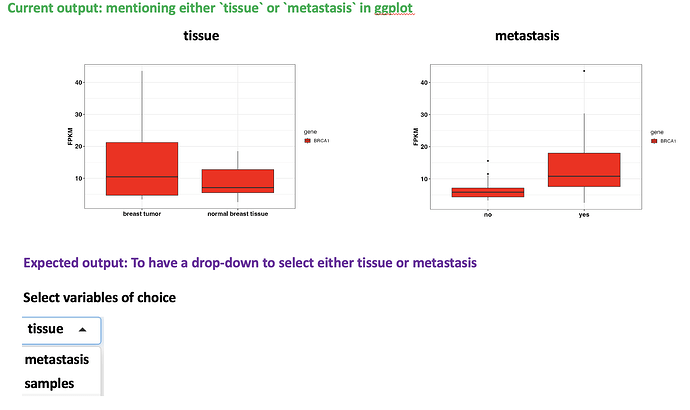
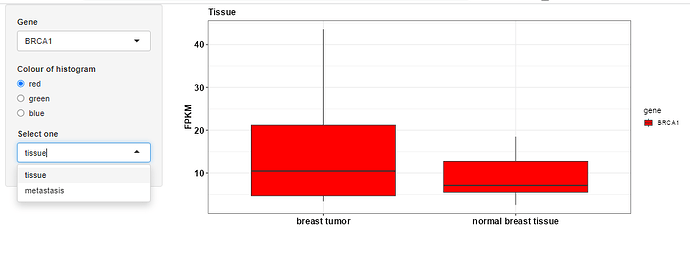
I have a question about adding a selection tab to list all available variables (columns) in R shiny to plot boxplot. At the moment I have to explicitly specify in the ggplot2 aesthetics the variable I am interested one by one (for instance, see the screenshot below: first for the tissue and then for the metastasis, etc., ). Having the selection tab in the way of drop-down like selecting the variables would be helpful.
# Data for plotting
dput(custom_data)
structure(list(gene = c("BRCA1", "BRCA1", "BRCA1", "BRCA1", "BRCA1",
"BRCA1", "BRCA1", "BRCA1", "BRCA1", "BRCA1", "BRCA1", "BRCA1",
"BRCA1", "BRCA1", "BRCA1", "BRCA1", "BRCA1", "BRCA1", "BRCA1",
"BRCA1", "BRCA2", "BRCA2", "BRCA2", "BRCA2", "BRCA2", "BRCA2",
"BRCA2", "BRCA2", "BRCA2", "BRCA2", "BRCA2", "BRCA2", "BRCA2",
"BRCA2", "BRCA2", "BRCA2", "BRCA2", "BRCA2", "BRCA2", "BRCA2",
"BRCA3", "BRCA3", "BRCA3", "BRCA3", "BRCA3", "BRCA3", "BRCA3",
"BRCA3", "BRCA3", "BRCA3", "BRCA1", "BRCA1", "BRCA1", "BRCA1",
"BRCA1", "BRCA1", "BRCA1", "BRCA1", "BRCA1", "BRCA1"), samples = c("CA.102548",
"CA.104338", "CA.105094", "CA.109745", "CA.1906415", "CA.1912627",
"CA.1924346", "CA.1926760", "CA.1927842", "CA.1933414", "CAP.102548",
"CAP.104338", "CAP.105094", "CAP.109745", "CAP.1906415", "CAP.1912627",
"CAP.1924346", "CAP.1926760", "CAP.1927842", "CAP.1933414", "CA.102548",
"CA.104338", "CA.105094", "CA.109745", "CA.1906415", "CA.1912627",
"CA.1924346", "CA.1926760", "CA.1927842", "CA.1933414", "CAP.102548",
"CAP.104338", "CAP.105094", "CAP.109745", "CAP.1906415", "CAP.1912627",
"CAP.1924346", "CAP.1926760", "CAP.1927842", "CAP.1933414", "CA.1940640",
"CA.2004407", "CA.2005288", "CA.2006047", "CA.2008260", "CA.2009329",
"CA.2009381", "CA.2009850", "CA.2017611", "CA.2039179", "CAP.1940640",
"CAP.2004407", "CAP.2005288", "CAP.2006047", "CAP.2008260", "CAP.2009329",
"CAP.2009381", "CAP.2009850", "CAP.2017611", "CAP.2039179"),
FPKM = c(30.37, 24.43, 43.58, 11.05, 3.38, 5.96, 4.26, 11.52,
9.87, 3.47, 15.91, 7.86, 8.58, 7.23, 5.43, 16.01, 3.29, 6.94,
9.86, 4.71, 0.38, 0.23, 0, 1.31, 0.53, 3.37, 1.21, 4.39,
3.91, 3.51, 9.28, 1.25, 2.61, 1.25, 0.31, 10.13, 0.54, 1.26,
6.39, 1.68, 11.28, 6.63, 5.46, 5.4, 14.64, 6.91, 9.49, 10,
3.85, 4.11, 17.85, 5.52, 6.32, 11.78, 15.59, 2.54, 6.19,
4.59, 18.49, 6.86), tissue = c("breast tumor", "breast tumor",
"breast tumor", "breast tumor", "breast tumor", "breast tumor",
"breast tumor", "breast tumor", "breast tumor", "breast tumor",
"normal breast tissue", "normal breast tissue", "normal breast tissue",
"normal breast tissue", "normal breast tissue", "normal breast tissue",
"normal breast tissue", "normal breast tissue", "normal breast tissue",
"normal breast tissue", "breast tumor", "breast tumor", "breast tumor",
"breast tumor", "breast tumor", "breast tumor", "breast tumor",
"breast tumor", "breast tumor", "breast tumor", "normal breast tissue",
"normal breast tissue", "normal breast tissue", "normal breast tissue",
"normal breast tissue", "normal breast tissue", "normal breast tissue",
"normal breast tissue", "normal breast tissue", "normal breast tissue",
"breast tumor", "breast tumor", "breast tumor", "breast tumor",
"breast tumor", "breast tumor", "breast tumor", "breast tumor",
"breast tumor", "breast tumor", "normal breast tissue", "normal breast tissue",
"normal breast tissue", "normal breast tissue", "normal breast tissue",
"normal breast tissue", "normal breast tissue", "normal breast tissue",
"normal breast tissue", "normal breast tissue"), metastasis = c("yes",
"yes", "yes", "no", "no", "yes", "no", "no", "yes", "no",
"yes", "yes", "yes", "no", "no", "yes", "no", "no", "yes",
"no", "yes", "yes", "yes", "no", "no", "yes", "no", "no",
"yes", "no", "yes", "yes", "yes", "no", "no", "yes", "no",
"no", "yes", "no", "yes", "no", "no", "yes", "no", "yes",
"no", "yes", "yes", "yes", "yes", "no", "no", "yes", "no",
"yes", "no", "yes", "yes", "yes")), class = "data.frame", row.names = c(NA,
-60L))
# Plot box plot
library(shiny)
library(ggplot2)
library(ggpubr)
ui <- fluidPage(
sidebarLayout(
sidebarPanel(
selectInput("thegene", "Gene", choices = c("BRCA1", "BRCA2", "BRCA3"), selected = "BRCA1"),
radioButtons("colour","Colour of histogram",choices=c("red","green","blue"),selected="red"),
width = 3
),
mainPanel(
plotOutput("boxplot"),
width = 9
)
)
)
server <- function(input, output, session) {
alldat <- reactive({
choices <- unique(custom_data$gene)
selected <- isolate(input$thegene)
if (!selected %in% choices) selected <- choices[1]
updateSelectInput(session, "thegene", choices = choices, selected = selected)
custom_data
})
dat <- reactive({
x <- alldat()
x[ x$gene == input$thegene,,drop=FALSE]
})
output$boxplot <- renderPlot({
ggplot(data = dat(), aes(x = tissue, y = FPKM, fill = gene)) +
geom_boxplot() +
theme_bw(base_size = 14) + xlab("") + ylab("FPKM") +
theme(axis.text=element_text(size=15, face = "bold", color = "black"),
axis.title=element_text(size=15, face = "bold", color = "black"),
strip.text = element_text(size=15, face = "bold", color = "black")) +
scale_fill_manual(values = input$colour)
})
}