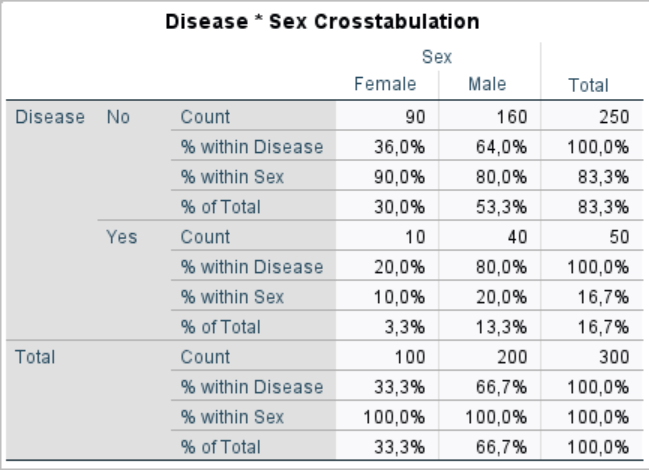
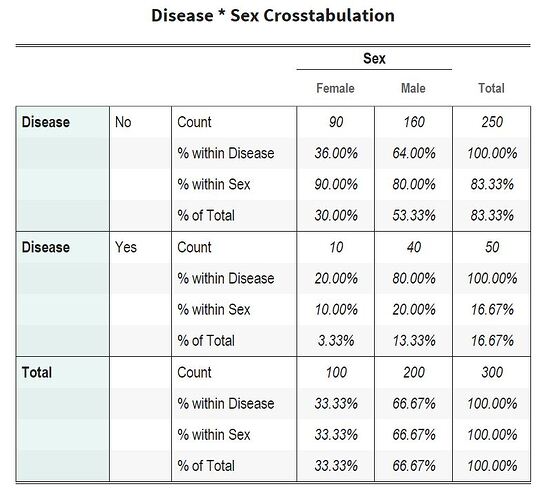
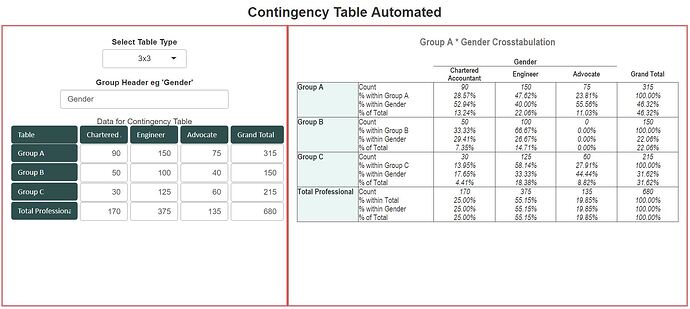
Hi Andrzej, finally done, fully automated for 2x2, 2x3, 3x2, and 3x3 contingency tables
Please note column and row headers having dark green backgrounds are all fully editable, it will reflect immediately on the Table. lengthier heading will be wrapped automatically. Fully automated, it has come very well , hope works for you as well
#######################################
# Loading Libraries
#######################################
library(shiny)
library(shinydashboard)
library(shinyjs)
library(stringr)
library(shinyWidgets)
library(dplyr)
library(kableExtra)
#######################################
# function for percentage with decimals
#######################################
fnpercent <- function(x, digits = 2, format = "f", ...) { # Create user-defined function
paste0(formatC(x * 100, format = format, digits = digits, ...), "%")
}
#######################################
# Dashboard UI
#######################################
ui <- fluidPage(
# Coloring textInput box #where you got:https://stackoverflow.com/questions/47478123/how-to-change-background-color-for-textInput-in-r-shiny
tags$head(tags$style(HTML('
#mRow0name, #mRow1name, #mRow2name, #mRow3name, #mRow4name,#mcol1name,#mcol2name,
#mcol3name,#mcol4name{
color:white; font-size:14px;
font-family: Calibri;
font-weight: normal;
background-color: #2f4f4f !important;
border: 2px double black;border-style:solid;
height: 32px;
overflow-y:none;
border: none;
}'))),
column(
width = 12,
align='center',
div(style = "margin-top:20px"),
HTML(paste('<h3><b>','Contingency Table Automated','</b><h5>')),
column(style = "border: 2px double red;height: 500px;overflow-y: auto;",
width = 5,
align = 'center',
div(style = "margin-top:20px"),
selectInput(
inputId = "mTableTypeChk",
label = 'Select Table Type',
choices = c('2x2','2x3','3x2','3x3'),
selected = '2x2',
width = '150px',
multiple = F
),
textInput(inputId = 'mgrptitle',label = "Group Header eg 'Gender'",value = 'Gender'),
HTML('Data for Contingency Table'),
splitLayout(cellWidths = c('25%','18%','18%','18%','20%'),
textInput(inputId = 'mRow0name',label = NULL,value = 'Table'),
textInput(inputId = 'mcol1name',label = NULL,value = 'Column1'),
textInput(inputId = 'mcol2name',label = NULL,value = 'Column2'),
textInput(inputId = 'mcol3name',label = NULL,value = 'Column3'),
textInput(inputId = 'mcol4name',label = NULL,value = 'Total')
),
div(style = "margin-top:-15px"),
splitLayout(cellWidths = c('25%','18%','18%','18%','20%'),
textInput(inputId = 'mRow1name',label = NULL,value = 'Row1'),
shinyWidgets::autonumericInput(inputId = "mValC11",label = NULL,value = 90,digitGroupSeparator = ",", decimalPlaces = 0),
shinyWidgets::autonumericInput(inputId = "mValC12",label = NULL,value = 160,digitGroupSeparator = ",", decimalPlaces = 0),
shinyWidgets::autonumericInput(inputId = "mValC13",label = NULL,value = 90,digitGroupSeparator = ",", decimalPlaces = 0),
shinyWidgets::autonumericInput(inputId = "mValC14",label = NULL,value = 90,digitGroupSeparator = ",", decimalPlaces = 0)
),
div(style = "margin-top:-15px"),
splitLayout(cellWidths = c('25%','18%','18%','18%','20%'),
textInput(inputId = 'mRow2name',label = NULL,value = 'Row2'),
shinyWidgets::autonumericInput(inputId = "mValC21",label = NULL,value = 10,digitGroupSeparator = ",", decimalPlaces = 0),
shinyWidgets::autonumericInput(inputId = "mValC22",label = NULL,value = 40,digitGroupSeparator = ",", decimalPlaces = 0),
shinyWidgets::autonumericInput(inputId = "mValC23",label = NULL,value = 10,digitGroupSeparator = ",", decimalPlaces = 0),
shinyWidgets::autonumericInput(inputId = "mValC24",label = NULL,value = 10,digitGroupSeparator = ",", decimalPlaces = 0)
),
div(style = "margin-top:-15px"),
splitLayout(cellWidths = c('25%','18%','18%','18%','20%'),
textInput(inputId = 'mRow3name',label = NULL,value = 'Row3'),
shinyWidgets::autonumericInput(inputId = "mValC31",label = NULL,value = 10,digitGroupSeparator = ",", decimalPlaces = 0),
shinyWidgets::autonumericInput(inputId = "mValC32",label = NULL,value = 40,digitGroupSeparator = ",", decimalPlaces = 0),
shinyWidgets::autonumericInput(inputId = "mValC33",label = NULL,value = 10,digitGroupSeparator = ",", decimalPlaces = 0),
shinyWidgets::autonumericInput(inputId = "mValC34",label = NULL,value = 10,digitGroupSeparator = ",", decimalPlaces = 0)
),
div(style = "margin-top:-15px"),
splitLayout(cellWidths = c('25%','18%','18%','18%','20%'),
textInput(inputId = 'mRow4name',label = NULL,value = 'Total'),
shinyWidgets::autonumericInput(inputId = "mValC41",label = NULL,value = 10,digitGroupSeparator = ",", decimalPlaces = 0),
shinyWidgets::autonumericInput(inputId = "mValC42",label = NULL,value = 40,digitGroupSeparator = ",", decimalPlaces = 0),
shinyWidgets::autonumericInput(inputId = "mValC43",label = NULL,value = 10,digitGroupSeparator = ",", decimalPlaces = 0),
shinyWidgets::autonumericInput(inputId = "mValC44",label = NULL,value = 10,digitGroupSeparator = ",", decimalPlaces = 0)
)
),
column(style = "border: 2px double red;height: 500px;overflow-y: auto;",
width = 7,
align = 'center',
htmlOutput('mshowmatrixtbl')
)
) #fluidrow closure
)
server <- function(input, output, session) {
#####################################################
# finally call function for contingency table
#####################################################
observeEvent(
c(input$mTableTypeChk,
input$mgrptitle,
input$mRow1name,
input$mRow2name,
input$mRow3name,
input$mRow4name,
input$mValC11,
input$mValC12,
input$mValC13,
input$mValC21,
input$mValC22,
input$mValC23,
input$mValC31,
input$mValC32,
input$mValC33,
input$mValC14,
input$mValC24,
input$mValC34,
input$mValC41,
input$mValC42,
input$mValC43,
input$mValC44),{
req(input$mgrptitle)
req(input$mValC11)
req(input$mValC12)
req(input$mValC13)
req(input$mValC21)
req(input$mValC22)
req(input$mValC23)
req(input$mValC31)
req(input$mValC32)
req(input$mValC33)
observeEvent(input$mTableTypeChk,{
if(input$mTableTypeChk=='2x2'){
shinyWidgets::updateAutonumericInput(session,inputId = 'mValC13',value = 0)
shinyWidgets::updateAutonumericInput(session,inputId = 'mValC23',value = 0)
shinyWidgets::updateAutonumericInput(session,inputId = 'mValC33',value = 0)
shinyWidgets::updateAutonumericInput(session,inputId = 'mValC31',value = 0)
shinyWidgets::updateAutonumericInput(session,inputId = 'mValC32',value = 0)
shinyWidgets::updateAutonumericInput(session,inputId = 'mValC33',value = 0)
}else if(input$mTableTypeChk=='2x3'){
shinyWidgets::updateAutonumericInput(session,inputId = 'mValC31',value = 0)
shinyWidgets::updateAutonumericInput(session,inputId = 'mValC32',value = 0)
shinyWidgets::updateAutonumericInput(session,inputId = 'mValC33',value = 0)
}else if(input$mTableTypeChk=='3x2'){
shinyWidgets::updateAutonumericInput(session,inputId = 'mValC13',value = 0)
shinyWidgets::updateAutonumericInput(session,inputId = 'mValC23',value = 0)
shinyWidgets::updateAutonumericInput(session,inputId = 'mValC33',value = 0)
}
})
shinyWidgets::updateAutonumericInput(session,inputId = 'mValC14',value = (input$mValC11+input$mValC12+input$mValC13))
shinyWidgets::updateAutonumericInput(session,inputId = 'mValC24',value = (input$mValC21+input$mValC22+input$mValC23))
shinyWidgets::updateAutonumericInput(session,inputId = 'mValC34',value = (input$mValC31+input$mValC32+input$mValC33))
shinyWidgets::updateAutonumericInput(session,inputId = 'mValC41',value = (input$mValC11+input$mValC21+input$mValC31))
shinyWidgets::updateAutonumericInput(session,inputId = 'mValC42',value = (input$mValC12+input$mValC22+input$mValC32))
shinyWidgets::updateAutonumericInput(session,inputId = 'mValC43',value = (input$mValC13+input$mValC23+input$mValC33))
shinyWidgets::updateAutonumericInput(session,inputId = 'mValC44',value = (input$mValC11+input$mValC21+input$mValC31+input$mValC12+input$mValC22+input$mValC32+input$mValC13+input$mValC23+input$mValC33))
df <- fnCreateTemporyTbl(mRow1name= input$mRow1name,mRow2name= input$mRow2name,mRow3name= input$mRow3name,mRow4name= input$mRow4name,mgrptitle= input$mgrptitle,mcol1name= input$mcol1name,mcol2name= input$mcol2name,mcol3name= input$mcol3name)
df <- data.frame(fnUpdateTemporyTbl(
df=df,
mValC11 = input$mValC11,mValC12 = input$mValC12,mValC13 = input$mValC13,
mValC21 = input$mValC21,mValC22 = input$mValC22,mValC23 = input$mValC23,
mValC31 = input$mValC31,mValC32 = input$mValC32,mValC33 = input$mValC33))
#########################################################################
# depending on your Contingency table type whether2x2 or 2x3 or 3x2, 3x3
# This code will delete unwanted coumn and rows as NA
# and then next two lines will delete ALL NA rows and ALL NA columns
#########################################################################
if(input$mTableTypeChk=='2x2'){
df[c(9:12),] <- NA
df[,5] <- NA
}else if(input$mTableTypeChk=='2x3'){
df[c(9:12),] <- NA
}else if(input$mTableTypeChk=='3x2'){
df[,5] <- NA
}
df <-df[rowSums(is.na(df)) != ncol(df), ] # Drop empty rows
df <- df[ , colSums(is.na(df)) != nrow(df)] # Drop empty columns
output$mshowmatrixtbl <- renderUI({
if(substr(input$mTableTypeChk,3,3)=='2'){
HTML(fnGenerateKblTbl2columns(df=df,mRow1name= input$mRow1name, mcol1name= input$mcol1name,mcol2name= input$mcol2name,mcol4name= input$mcol4name,mgrptitle= input$mgrptitle))
}else{
HTML(fnGenerateKblTbl3columns(df=df,mRow1name= input$mRow1name, mcol1name= input$mcol1name,mcol2name= input$mcol2name,mcol3name= input$mcol3name,mcol4name= input$mcol4name,mgrptitle= input$mgrptitle))
}
})
})
#########################################
# Building temporary dataframe called df
#########################################
fnCreateTemporyTbl <- function(mRow1name,mRow2name,mRow3name,mRow4name,mgrptitle,mcol1name,mcol2name,mcol3name){
Category <- c(mRow1name,NA,NA,NA,mRow2name,NA,NA,NA,mRow3name,NA,NA,NA,mRow4name,NA,NA,NA)
Subcategory <- c("Count",paste(" % within",mRow1name),paste("% within",mgrptitle),"% of Total",
"Count",paste("% within",mRow2name),paste("% within",mgrptitle),"% of Total",
"Count",paste("% within",mRow3name),paste("% within",mgrptitle),"% of Total",
"Count",paste("% within",'Total'),paste("% within",mgrptitle),"% of Total")
df <- data.frame(Category,Subcategory)
df[mcol1name] <- as.character()
df[mcol2name] <- as.character()
df[mcol3name] <- as.character()
df['Total'] <- as.character()
return(df)
}
#######################################
# function to update df
#######################################
fnUpdateTemporyTbl <- function(df,mValC11,mValC12,mValC13,mValC21,mValC22,mValC23,mValC31,mValC32,mValC33){
mTotline16 <- (mValC11 + mValC12 + mValC13)
mTotline56 <- (mValC21 + mValC22 + mValC23)
mTotline96 <- (mValC31 + mValC32 + mValC33)
mTotline133 <- (mValC11 + mValC21 + mValC31)
mTotline134 <- (mValC12 + mValC22 + mValC32)
mTotline135 <- (mValC13 + mValC23 + mValC33)
mTotline136 <- (mTotline133 + mTotline134 + mTotline135)
df[1,3] <- mValC11
df[1,4] <- mValC12
df[1,5] <- mValC13
df[1,6] <- mTotline16
df[5,3] <- mValC21
df[5,4] <- mValC22
df[5,5] <- mValC23
df[5,6] <- mTotline56
df[9,3] <- mValC31
df[9,4] <- mValC32
df[9,5] <- mValC33
df[9,6] <- mTotline96
df[13,3] <- mTotline133
df[13,4] <- mTotline134
df[13,5] <- mTotline135
df[13,6] <- mTotline136
# % of Disease
df[2,3] <- fnpercent(mValC11/mTotline16,2)
df[2,4] <- fnpercent(mValC12/mTotline16,2)
df[2,5] <- fnpercent(mValC13/mTotline16,2)
df[2,6] <- fnpercent(mTotline16/mTotline16,2)
df[6,3] <- fnpercent(mValC21/mTotline56,2)
df[6,4] <- fnpercent(mValC22/mTotline56,2)
df[6,5] <- fnpercent(mValC23/mTotline56,2)
df[6,6] <- fnpercent(mTotline56/mTotline56,2)
df[10,3] <- fnpercent(mValC31/mTotline96,2)
df[10,4] <- fnpercent(mValC32/mTotline96,2)
df[10,5] <- fnpercent(mValC33/mTotline96,2)
df[10,6] <- fnpercent(mTotline96/mTotline96,2)
df[14,3] <- fnpercent(mTotline133/mTotline136,2)
df[14,4] <- fnpercent(mTotline134/mTotline136,2)
df[14,5] <- fnpercent(mTotline135/mTotline136,2)
df[14,6] <- fnpercent(mTotline136/mTotline136,2)
# % of Sex
df[3,3] <- fnpercent(mValC11/mTotline133,2)
df[3,4] <- fnpercent(mValC12/mTotline134,2)
df[3,5] <- fnpercent(mValC13/mTotline135,2)
df[3,6] <- fnpercent(mTotline16/mTotline136,2)
df[7,3] <- fnpercent(mValC21/mTotline133,2)
df[7,4] <- fnpercent(mValC22/mTotline134,2)
df[7,5] <- fnpercent(mValC23/mTotline135,2)
df[7,6] <- fnpercent(mTotline56/mTotline136,2)
df[11,3] <- fnpercent(mValC31/mTotline133,2)
df[11,4] <- fnpercent(mValC32/mTotline134,2)
df[11,5] <- fnpercent(mValC33/mTotline135,2)
df[11,6] <- fnpercent(mTotline96/mTotline136,2)
df[15,3] <- fnpercent(mTotline133/mTotline136,2)
df[15,4] <- fnpercent(mTotline134/mTotline136,2)
df[15,5] <- fnpercent(mTotline135/mTotline136,2)
df[15,6] <- fnpercent(mTotline136/mTotline136,2)
# % of Total
df[4,3] <- fnpercent(mValC11/mTotline136,2)
df[4,4] <- fnpercent(mValC12/mTotline136,2)
df[4,5] <- fnpercent(mValC13/mTotline136,2)
df[4,6] <- fnpercent(mTotline16/mTotline136,2)
df[8,3] <- fnpercent(mValC21/mTotline136,2)
df[8,4] <- fnpercent(mValC22/mTotline136,2)
df[8,5] <- fnpercent(mValC23/mTotline136,2)
df[8,6] <- fnpercent(mTotline56/mTotline136,2)
df[12,3] <- fnpercent(mValC31/mTotline136,2)
df[12,4] <- fnpercent(mValC32/mTotline136,2)
df[12,5] <- fnpercent(mValC33/mTotline136,2)
df[12,6] <- fnpercent(mTotline96/mTotline136,2)
df[16,3] <- fnpercent(mTotline133/mTotline136,2)
df[16,4] <- fnpercent(mTotline134/mTotline136,2)
df[16,5] <- fnpercent(mTotline135/mTotline136,2)
df[16,6] <- fnpercent(mTotline136/mTotline136,2)
df <- sapply(df, as.character) # Convert all columns to character
df[is.na(df)] <- "" # Replace NA with blank
return(df)
}
#####################################################
# function to generate kbl table
#####################################################
our_strwrap <- function(x) lapply(strwrap(x, width = 15, simplify= FALSE), paste, collapse = "\n")
fnGenerateKblTbl2columns <- function(df,mRow1name,mcol1name,mcol2name,mcol4name,mgrptitle){
req(input$mgrptitle)
# Set a Title for the table
mtitle <- paste(mRow1name," * ",mgrptitle, 'Crosstabulation')
mtitle <- paste('<h4><STRONG><CENTER>',mtitle, '</CENTER></STRONG><h5>')
# Set a named vector for dynamic header
myGrpHeader <- c(" " = 2, mgrptitle = 2, " " = 1) #where you got this: https://stackoverflow.com/questions/45206908/kableextra-dynamic-add-header-above-labeling
# set vector names
names(myGrpHeader) <- c(" ", mgrptitle," ")
mBorderLineRow <- c(4,8)
if(substr(input$mTableTypeChk,1,1)=='3'){
mBorderLineRow <- c(mBorderLineRow,12)
}
n <- nrow(df)
#where you got this https://stackoverflow.com/questions/70871438/creating-horizontal-lines-after-each-collapsed-row-in-kableextra-table
df %>%
kbl(align = "llcccc",caption =HTML(mtitle),linesep = "\\addlinespace",row.names=FALSE,
escape=T, col.names = c("", "", our_strwrap(mcol1name),our_strwrap(mcol2name),our_strwrap(mcol4name))) %>%
kable_paper(c("striped"),full_width = F) %>%
column_spec(1, bold = T, background = "#91D1C233",border_left = T) %>%
column_spec(1,width = "8em", border_right = T) %>%
column_spec(2,width = "10em", border_right = T) %>%
column_spec(c(3,4,5), width = "8em") %>%
column_spec(c(3,4,5), italic = T,border_right = T,width = "8em") %>%
row_spec(0, bold = T, font_size = 13, extra_css = "border-bottom: 1px solid;") %>%
row_spec(mBorderLineRow, extra_css = "border-bottom: 1px solid;") %>%
row_spec(c(n), extra_css = "border-bottom: 1px solid;") %>%
# kable_styling(bootstrap_options = "striped") %>%
add_header_above(header = myGrpHeader,
bold = T,extra_css ="border-bottom: 1px solid;") %>%
kable_classic(lightable_options = "basic",
html_font = "\"Arial Narrow\", \"Source Sans Pro\", sans-serif")
}
fnGenerateKblTbl3columns <- function(df,mRow1name,mcol1name,mcol2name,mcol3name,mcol4name,mgrptitle){
req(input$mgrptitle)
# Set a Title for the table
mtitle <- paste(mRow1name," * ",mgrptitle, 'Crosstabulation')
mtitle <- paste('<h4><STRONG><CENTER>',mtitle, '</CENTER></STRONG><h5>')
# Set a named vector for dynamic header
myGrpHeader <- c(" " = 2, mgrptitle = 3, " " = 1) #where you got this: https://stackoverflow.com/questions/45206908/kableextra-dynamic-add-header-above-labeling
# set vector names
names(myGrpHeader) <- c(" ", mgrptitle," ")
mBorderLineRow <- c(4,8)
if(substr(input$mTableTypeChk,1,1)=='3'){
mBorderLineRow <- c(mBorderLineRow,12)
}
n <- nrow(df)
#where you got this https://stackoverflow.com/questions/70871438/creating-horizontal-lines-after-each-collapsed-row-in-kableextra-table
df %>%
kbl(align = "llcccc",caption =HTML(mtitle),linesep = "\\addlinespace",row.names=FALSE,
escape=T, col.names = c("", "", mcol1name,mcol2name,mcol3name,our_strwrap(mcol4name))) %>%
kable_paper(c("striped"),full_width = F) %>%
column_spec(1, bold = T, background = "#91D1C233",border_left = T) %>%
column_spec(1,width = "8em", border_right = T) %>%
column_spec(2,width = "10em", border_right = T) %>%
column_spec(c(3,4,5,6), width = "8em") %>%
column_spec(c(3,4,5,6), italic = T,border_right = T,width = "8em") %>%
row_spec(0, bold = T, font_size = 13, extra_css = "border-bottom: 1px solid;") %>%
row_spec(mBorderLineRow, extra_css = "border-bottom: 1px solid;") %>%
row_spec(c(n), extra_css = "border-bottom: 1px solid;") %>%
# kable_styling(bootstrap_options = "striped") %>%
add_header_above(header = myGrpHeader,
bold = T,extra_css ="border-bottom: 1px solid;") %>%
kable_classic(lightable_options = "basic",
html_font = "\"Arial Narrow\", \"Source Sans Pro\", sans-serif")
}
}
shinyApp(ui, server)