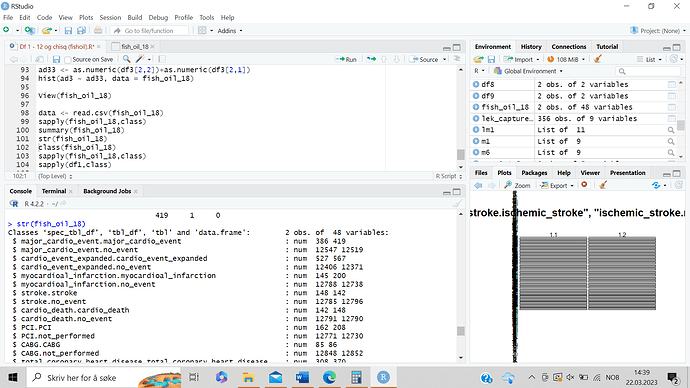
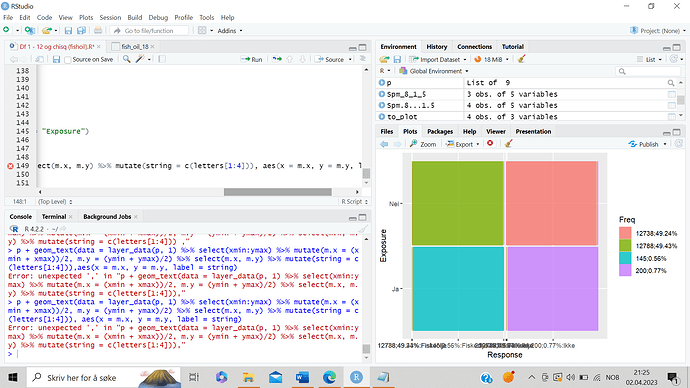
p + geom_text(data = layer_data(p, 1) %>% select(xmin:ymax) %>% mutate(m.x = (xmin + xmax))/2, m.y = (ymin + ymax)/2) %>% select(m.x, m.y) %>% mutate(string = c(letters[1:4])), aes(x = m.x, y = m.y, label = string)
Ok. This is the code, the last code. Everything goes like this:
library(ggmosaic)
install.packages("reprex")
library(reprex)
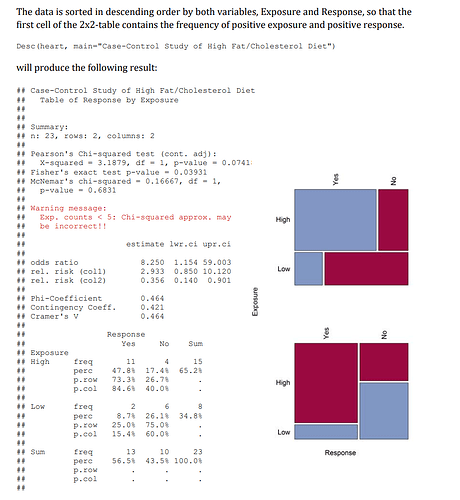
infarct2 <- as.table(matrix(c(145, 200, 12788, 12738), 2,2))
dimnames(infarct2) <- list(Exposure = c("Fiskeolje", "Ikke"), Response = c("Ja", "Nei"))
percentages <- round(100*prop.table(infarct2), 2)
etiquettes <- as.table(matrix(paste0(infarct2, ";", percentages, "%"), 2, 2))
dimnames(etiquettes) <- dimnames(infarct2)
to_plot <- as.data.frame(etiquettes)
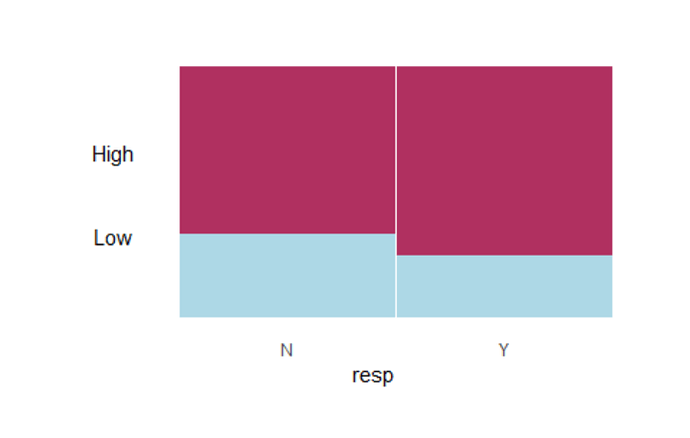
ggplot(data = to_plot) + geom_mosaic(aes(x = product(Response,Exposure), fill=Freq), na.rm = TRUE) + labs(x = "Response", y = "Exposure")
p <- ggplot(infarct2 %>% as_tibble()) + geom_mosaic(aes(weight = n, x = product(Response), fill = Exposure))
p + geom_text(data = layer_data(p, 1) %>% select(xmin:ymax) %>% mutate(m.x = (xmin + xmax))/2, m.y = (ymin + ymax)/2) %>% select(m.x, m.y) %>% mutate(string = c(letters[1:4])), aes(x = m.x, y = m.y, label = string)