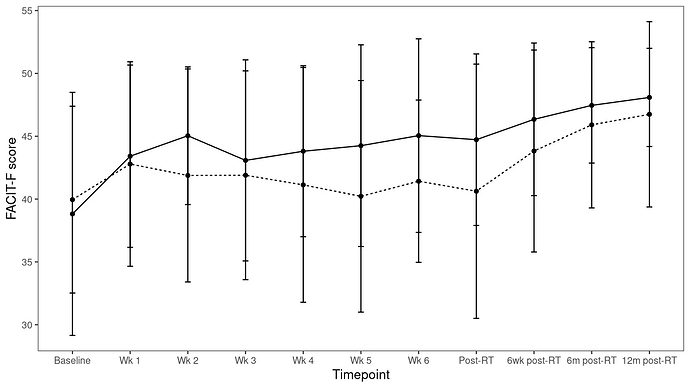
Hi, I have the below figure.
The code that produces this is:
linegraphFACITF_ALL <- ggplot(data=pergroupFACITF_ALL, aes(x=Timepoint, y=mean, group=Group)) +
geom_line(aes(linetype=Group)) +
geom_point() + geom_errorbar(aes(ymin=mean-sd, ymax=mean+sd), width=.1) +
labs(y = expression("FACIT-F score"), x="Timepoint") + theme_apa(legend.pos = "none")
I got two groups ('Exercise' and 'Usual Care' in the variable 'Group'). How can I make the error bars of the group that has the straight line be only above their points, and the error bars of the group with the dashed line be only below their points? I don't want to use position=dodge, as it still makes the figure cluttered.
Below is the dataframe 'pergroupFACITF_ALL' in csv format
Group,Timepoint,count,mean,sd
Exercise,Baseline,38,38.819047619047616,9.670500543696773
Exercise,Wk 1,38,43.41203703703704,7.251763453963447
Exercise,Wk 2,38,45.04054054054054,5.477705291018791
Exercise,Wk 3,38,43.078828828828826,7.9990341321341445
Exercise,Wk 4,38,43.806306306306304,6.8020414411974714
Exercise,Wk 5,38,44.25,8.023254038355303
Exercise,Wk 6,38,45.05,7.701503612590002
Exercise,Post-RT,38,44.726124885215796,6.822812537639399
Exercise,6wk post-RT,38,46.345588235294116,6.071547748946408
Exercise,6m post-RT,38,47.457142857142856,4.5911862951353495
Exercise,12m post-RT,38,48.08888888888889,3.906581070626291
Usual Care,Baseline,42,39.95614035087719,7.430754836032656
Usual Care,Wk 1,42,42.78846153846154,8.136105683894794
Usual Care,Wk 2,42,41.88095238095238,8.47489803076258
Usual Care,Wk 3,42,41.89484126984127,8.307297937697957
Usual Care,Wk 4,42,41.12916666666667,9.337948149407644
Usual Care,Wk 5,42,40.21875,9.213364679077749
Usual Care,Wk 6,42,41.42261904761905,6.456996494040687
Usual Care,Post-RT,42,40.625,10.116469182375278
Usual Care,6wk post-RT,42,43.824786324786324,8.035461310748326
Usual Care,6m post-RT,42,45.90625,6.610643299914894
Usual Care,12m post-RT,42,46.744360902255636,7.3735106797643954