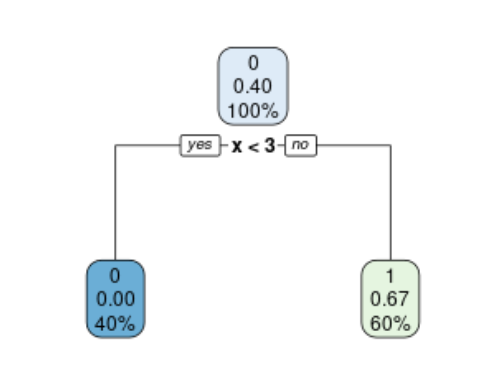
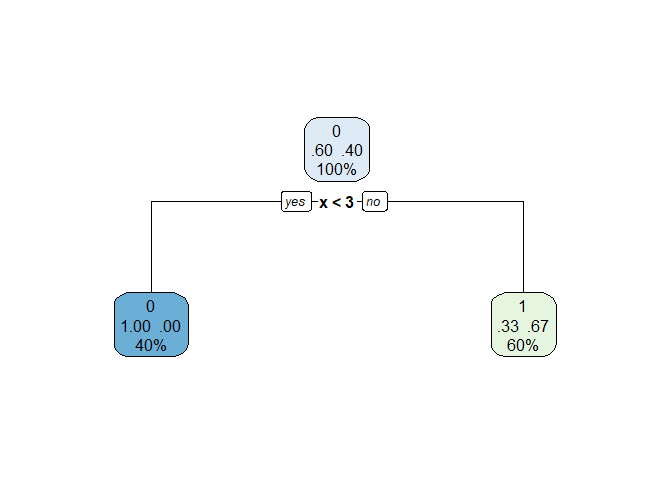
The cp table components can be extracted from the o object following
library(rpart)
library(rpart.plot)
df_ <- data.frame(x=c(1, 2, 3, 3, 3), y=c(0, 0, 1, 0, 1))
model<-rpart(y ~ x, data = df_, method="class", minbucket = 1, minsplit=1, xval=5)
(o <- summary(model)) |> str()
#> Call:
#> rpart(formula = y ~ x, data = df_, method = "class", minbucket = 1,
#> minsplit = 1, xval = 5)
#> n= 5
#>
#> CP nsplit rel error xerror xstd
#> 1 0.50 0 1.0 1 0.5477226
#> 2 0.01 1 0.5 2 0.4472136
#>
#> Variable importance
#> x
#> 100
#>
#> Node number 1: 5 observations, complexity param=0.5
#> predicted class=0 expected loss=0.4 P(node) =1
#> class counts: 3 2
#> probabilities: 0.600 0.400
#> left son=2 (2 obs) right son=3 (3 obs)
#> Primary splits:
#> x < 2.5 to the left, improve=1.066667, (0 missing)
#>
#> Node number 2: 2 observations
#> predicted class=0 expected loss=0 P(node) =0.4
#> class counts: 2 0
#> probabilities: 1.000 0.000
#>
#> Node number 3: 3 observations
#> predicted class=1 expected loss=0.3333333 P(node) =0.6
#> class counts: 1 2
#> probabilities: 0.333 0.667
#>
#> List of 14
#> $ frame :'data.frame': 3 obs. of 9 variables:
#> ..$ var : chr [1:3] "x" "<leaf>" "<leaf>"
#> ..$ n : int [1:3] 5 2 3
#> ..$ wt : num [1:3] 5 2 3
#> ..$ dev : num [1:3] 2 0 1
#> ..$ yval : num [1:3] 1 1 2
#> ..$ complexity: num [1:3] 0.5 0.01 0.01
#> ..$ ncompete : int [1:3] 0 0 0
#> ..$ nsurrogate: int [1:3] 0 0 0
#> ..$ yval2 : num [1:3, 1:6] 1 1 2 3 2 1 2 0 2 0.6 ...
#> .. ..- attr(*, "dimnames")=List of 2
#> .. .. ..$ : NULL
#> .. .. ..$ : chr [1:6] "" "" "" "" ...
#> $ where : Named int [1:5] 2 2 3 3 3
#> ..- attr(*, "names")= chr [1:5] "1" "2" "3" "4" ...
#> $ call : language rpart(formula = y ~ x, data = df_, method = "class", minbucket = 1, minsplit = 1, xval = 5)
#> $ terms :Classes 'terms', 'formula' language y ~ x
#> .. ..- attr(*, "variables")= language list(y, x)
#> .. ..- attr(*, "factors")= int [1:2, 1] 0 1
#> .. .. ..- attr(*, "dimnames")=List of 2
#> .. .. .. ..$ : chr [1:2] "y" "x"
#> .. .. .. ..$ : chr "x"
#> .. ..- attr(*, "term.labels")= chr "x"
#> .. ..- attr(*, "order")= int 1
#> .. ..- attr(*, "intercept")= int 1
#> .. ..- attr(*, "response")= int 1
#> .. ..- attr(*, ".Environment")=<environment: R_GlobalEnv>
#> .. ..- attr(*, "predvars")= language list(y, x)
#> .. ..- attr(*, "dataClasses")= Named chr [1:2] "numeric" "numeric"
#> .. .. ..- attr(*, "names")= chr [1:2] "y" "x"
#> $ cptable : num [1:2, 1:5] 0.5 0.01 0 1 1 ...
#> ..- attr(*, "dimnames")=List of 2
#> .. ..$ : chr [1:2] "1" "2"
#> .. ..$ : chr [1:5] "CP" "nsplit" "rel error" "xerror" ...
#> $ method : chr "class"
#> $ parms :List of 3
#> ..$ prior: num [1:2(1d)] 0.6 0.4
#> .. ..- attr(*, "dimnames")=List of 1
#> .. .. ..$ : chr [1:2] "1" "2"
#> ..$ loss : num [1:2, 1:2] 0 1 1 0
#> ..$ split: num 1
#> $ control :List of 9
#> ..$ minsplit : num 1
#> ..$ minbucket : num 1
#> ..$ cp : num 0.01
#> ..$ maxcompete : int 4
#> ..$ maxsurrogate : int 5
#> ..$ usesurrogate : int 2
#> ..$ surrogatestyle: int 0
#> ..$ maxdepth : int 30
#> ..$ xval : num 5
#> $ functions :List of 3
#> ..$ summary:function (yval, dev, wt, ylevel, digits)
#> ..$ print :function (yval, ylevel, digits, nsmall)
#> ..$ text :function (yval, dev, wt, ylevel, digits, n, use.n)
#> $ numresp : int 4
#> $ splits : num [1, 1:5] 5 -1 1.07 2.5 0
#> ..- attr(*, "dimnames")=List of 2
#> .. ..$ : chr "x"
#> .. ..$ : chr [1:5] "count" "ncat" "improve" "index" ...
#> $ variable.importance: Named num 1.07
#> ..- attr(*, "names")= chr "x"
#> $ y : int [1:5] 1 1 2 1 2
#> $ ordered : Named logi FALSE
#> ..- attr(*, "names")= chr "x"
#> - attr(*, "xlevels")= Named list()
#> - attr(*, "ylevels")= chr [1:2] "0" "1"
#> - attr(*, "class")= chr "rpart"
Created on 2023-03-09 with reprex v2.0.2