Rick@starz and I (Richard Careaga) invite discussion on when lm() should be avoided for binary values if at all? What alternative techniques should be considered?
- Continuous against continuous: both sides of the
lm()formula are continuous variables - Continuous against binary: left-hand side (LHS) continuous response with a binary predictor (right hand side RHS)
- Binary against continuous: A LHS binary response with a RHS continuous predictor
- Binary against binary: Both LHS and RHS are binary.
As a motivating example, mtcars includes four variables mpg, drat, vs and am
From help(mtcars)
mpg Miles/(US) gallon
drat Rear axle ratio
vs Engine (0 = V-shaped, 1 = straight)
am Transmission (0 = automatic, 1 = manual)
LHS continuous and RHS continuous
In the usual case of ordinary least squares linear regression both the LHS response variable and the RHS predictor variable are continuous
require(ggplot2)
#> Loading required package: ggplot2
mtcars |> ggplot(aes(drat,mpg)) +
geom_point() +
geom_smooth(method = "lm") +
theme_minimal()
#> `geom_smooth()` using formula = 'y ~ x'
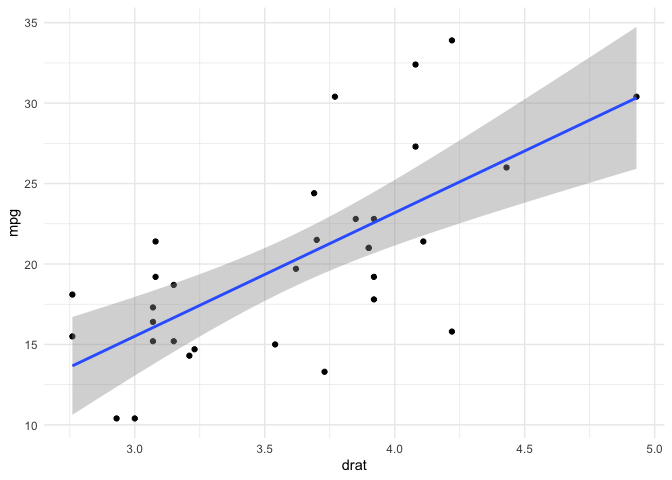
(fit <- lm(mpg ~ drat, data = mtcars))
#>
#> Call:
#> lm(formula = mpg ~ drat, data = mtcars)
#>
#> Coefficients:
#> (Intercept) drat
#> -7.525 7.678
summary(fit)
#>
#> Call:
#> lm(formula = mpg ~ drat, data = mtcars)
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -9.0775 -2.6803 -0.2095 2.2976 9.0225
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) -7.525 5.477 -1.374 0.18
#> drat 7.678 1.507 5.096 1.78e-05 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Residual standard error: 4.485 on 30 degrees of freedom
#> Multiple R-squared: 0.464, Adjusted R-squared: 0.4461
#> F-statistic: 25.97 on 1 and 30 DF, p-value: 1.776e-05
par(mfrow = c(2,2))
plot(fit)
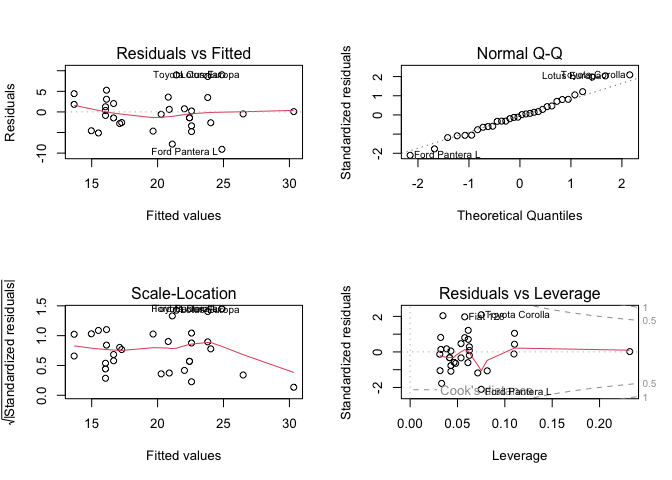
LHS continuous and RHS binary
require(ggplot2)
#> Loading required package: ggplot2
mtcars |> ggplot(aes(vs,drat)) +
geom_point() +
geom_smooth(method = "lm") +
theme_minimal()
#> `geom_smooth()` using formula = 'y ~ x'
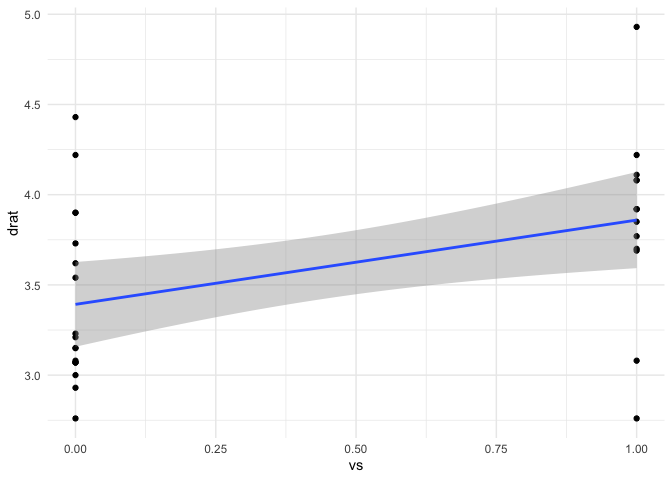
(fit2 <- lm(drat ~ vs, data = mtcars))
#>
#> Call:
#> lm(formula = drat ~ vs, data = mtcars)
#>
#> Coefficients:
#> (Intercept) vs
#> 3.3922 0.4671
summary(fit2)
#>
#> Call:
#> lm(formula = drat ~ vs, data = mtcars)
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -1.09929 -0.31472 -0.04929 0.23351 1.07071
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 3.3922 0.1150 29.492 <2e-16 ***
#> vs 0.4671 0.1739 2.686 0.0117 *
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Residual standard error: 0.488 on 30 degrees of freedom
#> Multiple R-squared: 0.1938, Adjusted R-squared: 0.167
#> F-statistic: 7.214 on 1 and 30 DF, p-value: 0.01168
par(mfrow = c(2,2))
plot(fit2)
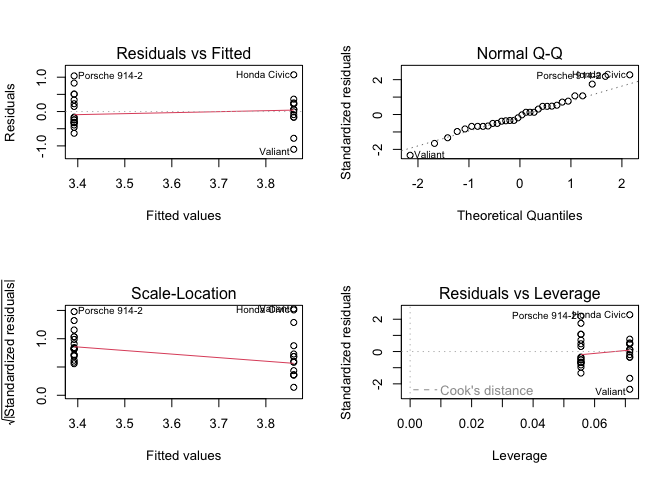
RHS binary and RHS continuous
require(ggplot2)
#> Loading required package: ggplot2
mtcars |> ggplot(aes(drat,vs)) +
geom_point() +
geom_smooth(method = "lm") +
theme_minimal()
#> `geom_smooth()` using formula = 'y ~ x'
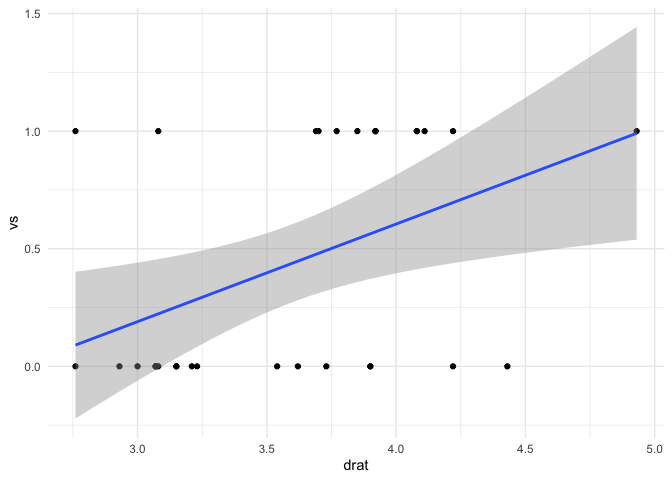
(fit3 <- lm(vs ~ drat, data = mtcars))
#>
#> Call:
#> lm(formula = vs ~ drat, data = mtcars)
#>
#> Coefficients:
#> (Intercept) drat
#> -1.055 0.415
summary(fit3)
#>
#> Call:
#> lm(formula = vs ~ drat, data = mtcars)
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -0.7834 -0.2791 -0.1754 0.4283 0.9097
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) -1.0552 0.5617 -1.879 0.0700 .
#> drat 0.4150 0.1545 2.686 0.0117 *
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Residual standard error: 0.46 on 30 degrees of freedom
#> Multiple R-squared: 0.1938, Adjusted R-squared: 0.167
#> F-statistic: 7.214 on 1 and 30 DF, p-value: 0.01168
par(mfrow = c(2,2))
plot(fit3)
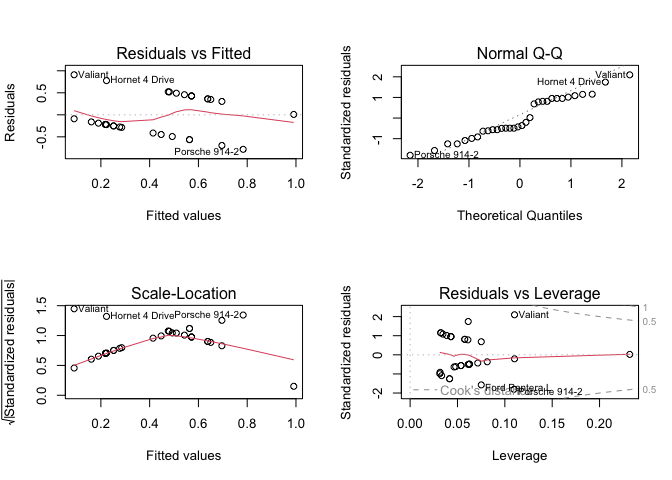
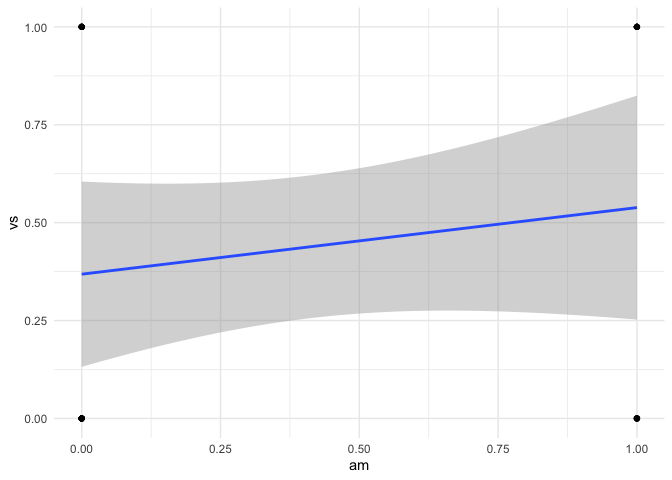
LHS binary and RHS binary
require(ggplot2)
#> Loading required package: ggplot2
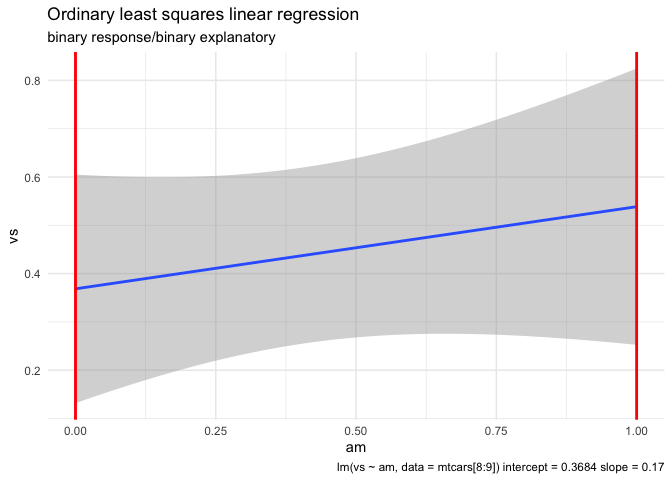
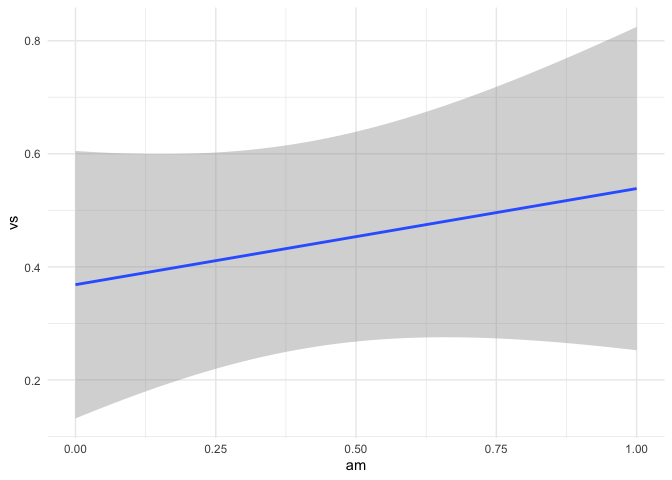
mtcars |> ggplot(aes(am,vs)) +
geom_point() +
geom_smooth(method = "lm") +
theme_minimal()
#> `geom_smooth()` using formula = 'y ~ x'
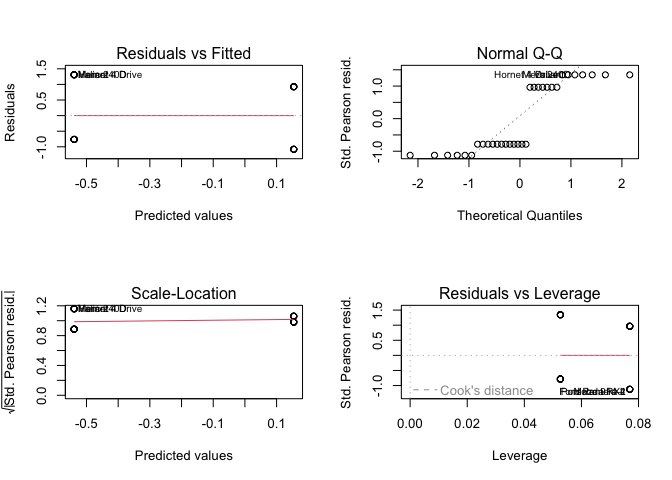
(fit4 <- lm(vs ~ am, data = mtcars))
#>
#> Call:
#> lm(formula = vs ~ am, data = mtcars)
#>
#> Coefficients:
#> (Intercept) am
#> 0.3684 0.1700
summary(fit4)
#>
#> Call:
#> lm(formula = vs ~ am, data = mtcars)
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -0.5385 -0.3684 -0.3684 0.4615 0.6316
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 0.3684 0.1159 3.180 0.00341 **
#> am 0.1700 0.1818 0.935 0.35704
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Residual standard error: 0.505 on 30 degrees of freedom
#> Multiple R-squared: 0.02834, Adjusted R-squared: -0.004049
#> F-statistic: 0.875 on 1 and 30 DF, p-value: 0.357
par(mfrow = c(2,2))
plot(fit4)

Created on 2023-04-09 with reprex v2.0.2