Hi everyone, Updating the post to provide some progress and to more fully detail what I think the logic should be. Basically, in the for loop, I want to do the following. First, for the first row in the dataframe, I want to check if the name exists in the name_group_id list we initialized. If It does, then assign the group_id associated with that name. If not, then check if the location exists in the location_group_id list. If it does, return that group_id, if it does not, then create a new group_id and add this row's name/group_id to the name_group_id and the location/group_id to the location_group_id
This code does not work yet, as row three should have group_id 1 because name interacted with location 1 in row 1. But it seems close. Help tweaking this code would be awesome.
Thanks, everyone.
# Create data frame with the sample data
df <- data.frame(name = c("a", "a", "b", "b", "b", "c", "c", "d", "d", "e", "e", "f", "g", "g", "h"),
location = c(1, 2, 1, 3, 4, 3, 2, 5, 6, 7, 8, 4, 9, 10, 5))
# Initialize two lists
name_id <- list()
location_id <- list()
# Counter for creating new group IDs
group_id_counter <- 0
# For loop that implements the logic
for (i in 1:nrow(df)) {
row <- df[i, ]
name <- row$_name
location <- row$location
# Check if the name exists in name_id
if (name %in% names(name_id)) {
group_id <- name_group_id[[name]]
} else {
# Check if the location exists in location_group_id
if (location %in% names(location _group_id)) {
group_id <- location _group_id[[location ]]
} else {
group_id_counter <- group_id_counter + 1
group_id <- paste0("Group ", group_id_counter)
name_id[[name]] <- group_id
location _group_id[[location ]] <- group_id
}
}
# Assign the group_id to the current row
df[i, "group_id"] <- group_id
}
OLD thinking below
I have what I think is a set partition problem requiring a for loop. My dataset that looks similar to this
group1 <- sample(c("person a" , "person b" , "person c",
"person d" , "person e" , "person f", "person g"),
25, replace = TRUE )
group2 <- sample(c("veggie" , "fruit" , "meat",
"dairy" , "dirt"),
25, replace = TRUE )
df <- data.frame(group1, group2)
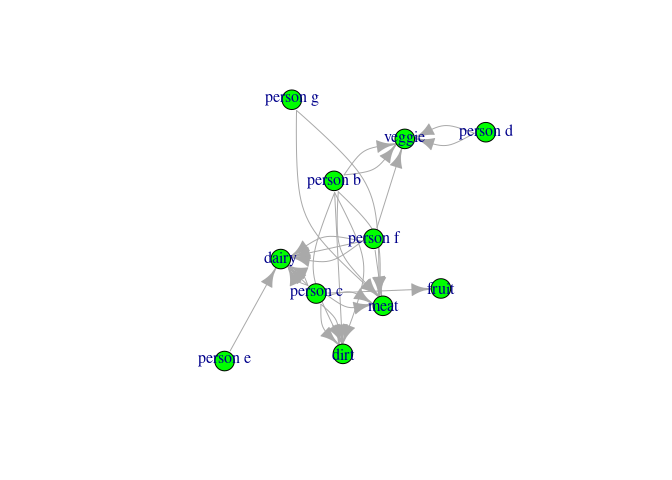
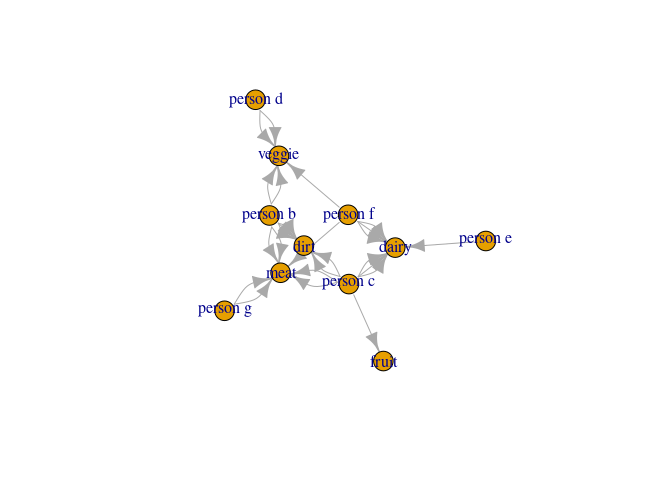
You could think of these data as foods (group2) people (group1) eat.
What I would like to do is determine the unique group memberships at play. So if person 1 eats dirt, but no other person does and person 1 eats nothing else, then person 1 is in ID 1.
If both person 2 and person 5 each fruit, but no other person does, then they could be group 2. But, crucially, any overlap creates a group, so if person 5 from above eats both fruits and veggies, and person three also eats veggies, then persons 2, 5, and 3 become a common ID (e.g. ID 2).
My real data has distinct combinations, but no repeat combinations.
My real data is ~7million rows long and the combinations are more complicated, but the general idea stands.
I have toyed with a few ways to try this but increasingly think I need to employ some sort of for loop, which I am not very skilled at.
Any help would be great. I also welcome pointers to other solutions, in case I missed any.
thanks!