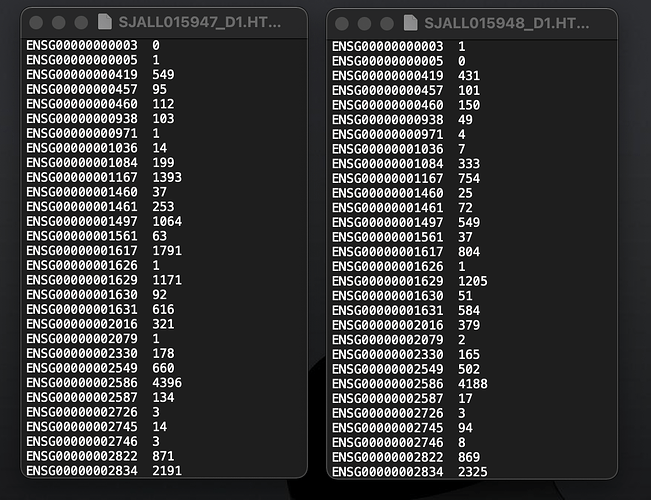
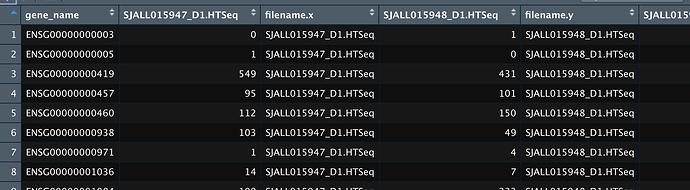
(This illustrates why screenshots are so iffy—I saw only the two columns.)
There are ways to select columns based on name patterns, but this can be done simply by taking advantage of the regular layout: we want to discard every third column. We can do this by using subsetting based on the column index.
dat <- data.frame(
A =c("a", "b", "c", "d", "e", "f", "g", "h", "i", "j", "k", "l", "m", "n", "o", "p", "q", "r", "s", "t", "u", "v", "w", "x", "y", "z"), B = 1:26,
C = c("A", "B", "C", "D", "E", "F", "G", "H", "I", "J", "K", "L", "M", "N", "O", "P", "Q", "R", "S", "T", "U", "V", "W", "X", "Y", "Z"),
D = c("a", "b", "c", "d", "e", "f", "g", "h", "i", "j", "k", "l", "m", "n", "o", "p", "q", "r", "s", "t", "u", "v", "w", "x", "y", "z"), E = 1:26,
F = c("A", "B", "C", "D", "E", "F", "G", "H", "I", "J", "K", "L", "M", "N", "O", "P", "Q", "R", "S", "T", "U", "V", "W", "X", "Y", "Z"),
G = c("a", "b", "c", "d", "e", "f", "g", "h", "i", "j", "k", "l", "m", "n", "o", "p", "q", "r", "s", "t", "u", "v", "w", "x", "y", "z"), H = 1:26,
I = c("A", "B", "C", "D", "E", "F", "G", "H", "I", "J", "K", "L", "M", "N", "O", "P", "Q", "R", "S", "T", "U", "V", "W", "X", "Y", "Z")
)
dat
#> A B C D E F G H I
#> 1 a 1 A a 1 A a 1 A
#> 2 b 2 B b 2 B b 2 B
#> 3 c 3 C c 3 C c 3 C
#> 4 d 4 D d 4 D d 4 D
#> 5 e 5 E e 5 E e 5 E
#> 6 f 6 F f 6 F f 6 F
#> 7 g 7 G g 7 G g 7 G
#> 8 h 8 H h 8 H h 8 H
#> 9 i 9 I i 9 I i 9 I
#> 10 j 10 J j 10 J j 10 J
#> 11 k 11 K k 11 K k 11 K
#> 12 l 12 L l 12 L l 12 L
#> 13 m 13 M m 13 M m 13 M
#> 14 n 14 N n 14 N n 14 N
#> 15 o 15 O o 15 O o 15 O
#> 16 p 16 P p 16 P p 16 P
#> 17 q 17 Q q 17 Q q 17 Q
#> 18 r 18 R r 18 R r 18 R
#> 19 s 19 S s 19 S s 19 S
#> 20 t 20 T t 20 T t 20 T
#> 21 u 21 U u 21 U u 21 U
#> 22 v 22 V v 22 V v 22 V
#> 23 w 23 W w 23 W w 23 W
#> 24 x 24 X x 24 X x 24 X
#> 25 y 25 Y y 25 Y y 25 Y
#> 26 z 26 Z z 26 Z z 26 Z
# pick out column index numbers that are not divisible by 3 leaving
# no remainder using modulus operator, because that's every third
# column
keepers <- which(1:9 %% 3 != 0)
dat[keepers]
#> A B D E G H
#> 1 a 1 a 1 a 1
#> 2 b 2 b 2 b 2
#> 3 c 3 c 3 c 3
#> 4 d 4 d 4 d 4
#> 5 e 5 e 5 e 5
#> 6 f 6 f 6 f 6
#> 7 g 7 g 7 g 7
#> 8 h 8 h 8 h 8
#> 9 i 9 i 9 i 9
#> 10 j 10 j 10 j 10
#> 11 k 11 k 11 k 11
#> 12 l 12 l 12 l 12
#> 13 m 13 m 13 m 13
#> 14 n 14 n 14 n 14
#> 15 o 15 o 15 o 15
#> 16 p 16 p 16 p 16
#> 17 q 17 q 17 q 17
#> 18 r 18 r 18 r 18
#> 19 s 19 s 19 s 19
#> 20 t 20 t 20 t 20
#> 21 u 21 u 21 u 21
#> 22 v 22 v 22 v 22
#> 23 w 23 w 23 w 23
#> 24 x 24 x 24 x 24
#> 25 y 25 y 25 y 25
#> 26 z 26 z 26 z 26