Project
RStudio Cloud project: Posit Cloud
Live app: https://kevinrue.shinyapps.io/isee-shiny-contest/
GitHub repository: GitHub - kevinrue/isee-shiny-contest: iSEE repository for the Shiny contest
Description
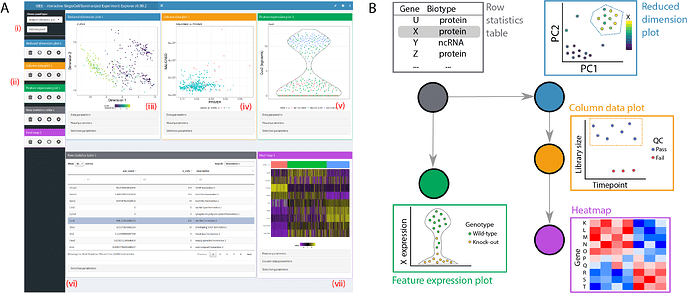
iSEE (interactive SummarizedExperiment Explorer) is designed for interactive exploration of high-throughput biological data sets stored in the SummarizedExperiment S4 class. It enables simultaneous visualisation of experimental data, associated metadata and analysis results, empowering users to discover new relationships in their data. iSEE combines shiny with other packages designed for interactive user interfaces (e.g., shinydashboard, shinjs, shinyAce, colourpicker, htmltools), with further extensions to deliver a full-featured application accessible to both novice users and experienced developers. We have demonstrated the capabilities of iSEE by applying it to a diverse range of real data sets (see links below).
Key features
-
A multi-panel layout to simultaneously visualise multiple aspects of the dataset.
-
Point selections using shiny brushes or lasso polygons drawn by consecutive clicks.
- Multiple concurrent selections supported in versions ≥ 1.3.8
-
Dynamic linking between plot and table panels, allowing users to transmit information across multiple panels via point selection.
-
Dynamic addition, removal, reordering, and resizing of panels.
-
Automatic tracking, storage, and rendering of the exact R code in a shinyAce editor to allow regeneration of all visible plots in a given application instance.
-
Intelligent density-dependent downsampling of data points shown for large data sets.
-
Custom colour scale specification for individual and groups of discrete and continuous variables.
-
Construction of bespoke interactive guided tours using rintrojs, which allow users to communicate their results through a step-by-step description of the salient features of the data.
-
Dynamic detection of discrete and continuous covariates, switching individual plot panels between scatter plots, violin plots, and “square” plots, with protection against factors including an excessive number of levels.
-
Ability for users to programmatically define additional custom panels based on any number of user-defined functions performing on-the-fly calculations.
-
Ability to preconfigure app instances, including point selections and state of collapsible panels, to launch individual app instances in any predefined state.
-
Retrieval of gene annotations on the fly by using either user-defined functions or the built-in
annotateEntrezorannotateEnsemblfunctions. -
Hands-free control of the app using voice recognition by annyang in versions ≥ 1.3.2.
Links
-
Source code for the iSEE package: GitHub - iSEE/iSEE: R/shiny interface for interactive visualization of data in SummarizedExperiment objects
-
Documentation and vignettes: Bioconductor - iSEE
-
Published article: https://f1000research.com/articles/7-741/v1
-
Direct links to the tours showcased in the article: https://marionilab.cruk.cam.ac.uk
-
Source code to construct the tours showcased in the article: GitHub - iSEE/iSEE2018: Code and manuscript files for the iSEE paper
-
Source code for minimal iSEE applications that each demonstrates a custom plot or table panel: GitHub - kevinrue/iSEE_custom: Examples of custom panels for the iSEE package
-
Source code to reproduce entire analyses as instances of iSEE apps for publicly available data sets: GitHub - iSEE/iSEE_instances: A gallery-repository to store the commands to reproduce entire analyses for iSEE instances on public/own data
-
A set of independent iSEE instances showcasing data from a recently published scientific study: Single-Cell Gene Expression in Regulatory T cells — Teichmann Lab
Technical details
-
The interface layout is built using the shinydashboard package, with colour-coded panel types representing visualisations of different aspects of the data.
-
Reactive observers are generated for every panel using
forloops and a set of predefined constant identifiers for every type of panel and input widget, combined with a numerical index to generate unique input identifiers. -
Panel parameters are stored in separate
DataFrames to enable the application state to be recovered at any time. -
Persistent lassos are created by handling click events in a context-dependent manner.
-
Transmission of selections between two or more panels is performed using a graph-based framework that can handle hierarchical dependencies.
-
Code tracking is achieved by memorizing the ggplot2 code that is to be evaluated upon the reactive evaluation of plot updates.
-
From the very start, we have continuously refactored the app functionality in the iSEE package, to deliver an application that be launched from as little as a single command for any dataset.
Contributors
Disclaimer
The demonstration app for the RStudio shiny contest uses the latest release version of the iSEE package on Bioconductor. A range of new exciting features are available from the the latest Bioconductor development version, and the very latest features (e.g., multiple selections, voice recognition, updated UI) may be obtained directly from the master branch of our GitHub repository.
We encourage you to try them out!