@Max I got a little lost trying to work through the example in the paper / blog I'd shared ![]() (https://saattrupdan.github.io/2020-03-01-bootstrap-prediction/ ) and may have messed-up trying to encode it... :-/
(https://saattrupdan.github.io/2020-03-01-bootstrap-prediction/ ) and may have messed-up trying to encode it... :-/
- I stuck with bootstrapping for the model variance component
- Used cross validation for the variance in the residuals (per your suggestion) so skipped the whole adjustment around 632 and training/validation error
Here is a quick (attempted) example with decision trees and 90% prediction interval:
library(tidyverse)
library(tidymodels)
set.seed(123)
iris <- as_tibble(iris)
split <- initial_split(iris)
train <- training(split)
test <- testing(split)
model_variance <- rsample::bootstraps(train, sqrt(nrow(test))) %>%
mutate(mod = map(splits, ~ rpart::rpart(Sepal.Length ~ Sepal.Width, data = analysis(.x))),
test_preds = map(mod, ~ tibble(
pred = predict(.x, test),
index = seq_len(nrow(test))
))) %>%
unnest(test_preds) %>%
group_by(index) %>%
mutate(m = pred - mean(pred)) %>%
select(index, m, pred)
sampling_variance <- rsample::vfold_cv(train, 10) %>%
mutate(
mod = map(splits, ~ rpart::rpart(Sepal.Length ~ Sepal.Width, data = analysis(.x))),
resids = map2(
mod,
splits,
~ predict(.x, assessment(.y)) - assessment(.y)$Sepal.Length
)
) %>%
select(resids) %>%
unnest(resids)
tidyr::crossing(model_variance, sampling_variance) %>%
mutate(c = m + resids + pred) %>%
group_by(index) %>%
summarise(q_05 = quantile(c, 0.05),
q_95 = quantile(c, 0.95)) %>%
bind_cols(test, rpart::rpart(Sepal.Length ~ Sepal.Width, data = train) %>% predict(test) %>% tibble(.pred = .)) %>%
ggplot(aes(x = Sepal.Width))+
geom_point(aes(y = Sepal.Length))+
geom_line(aes(y = q_05), colour = "red")+
geom_line(aes(y = q_95), colour = "red")+
geom_line(aes(y = .pred), colour = "blue")+
ylim(c(3, 9))
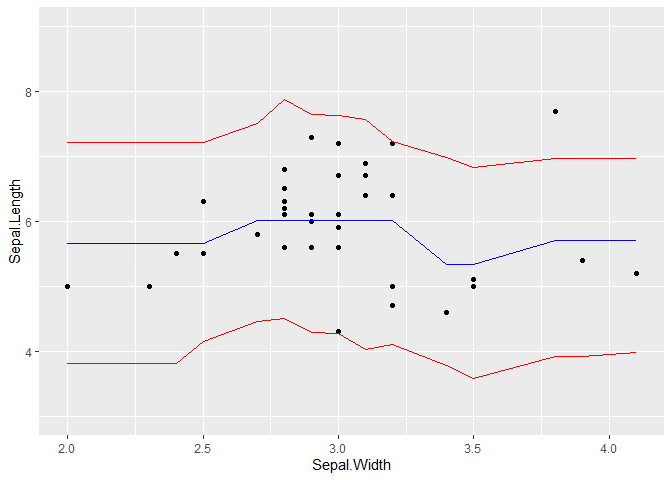
Created on 2021-02-12 by the reprex package (v0.3.0)
Could functionalize and edit so could take in a parsnip model specification and a recipe (but just wanted to get down a first attempt as wasn't sure if was doing correctly... ![]() ).
).
Other resources I found were this more simple approach:
Which essentially just uses randomized residuals (I imagine you would recommend using an alternative method for sampling the residuals here instead of pulling them from the model training/analysis set) -- not sure if this method may only be appropriate in linear models though...? Then there is the similar approach on Cross Validated which uses variance adjusted residuals (again assuming confined to linear models, and in this case relies on measure of influence).
