First of all, I thank you very much for watching my question
Please take a look at a dynamic diagram first
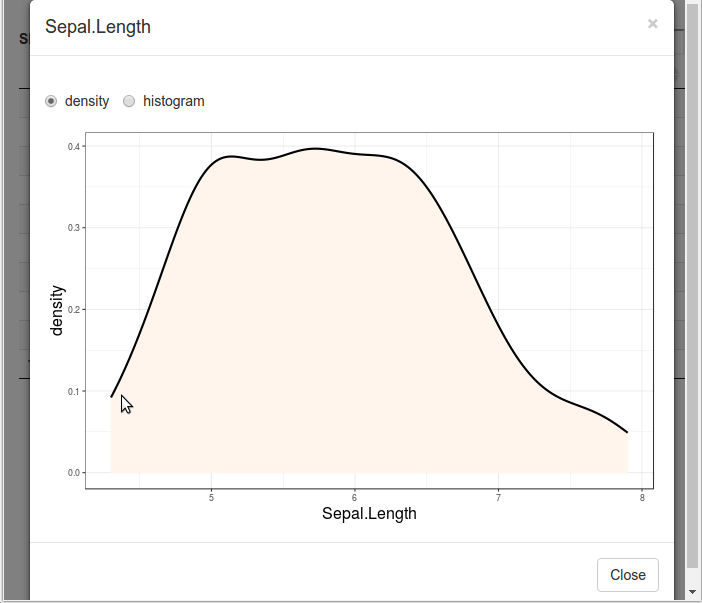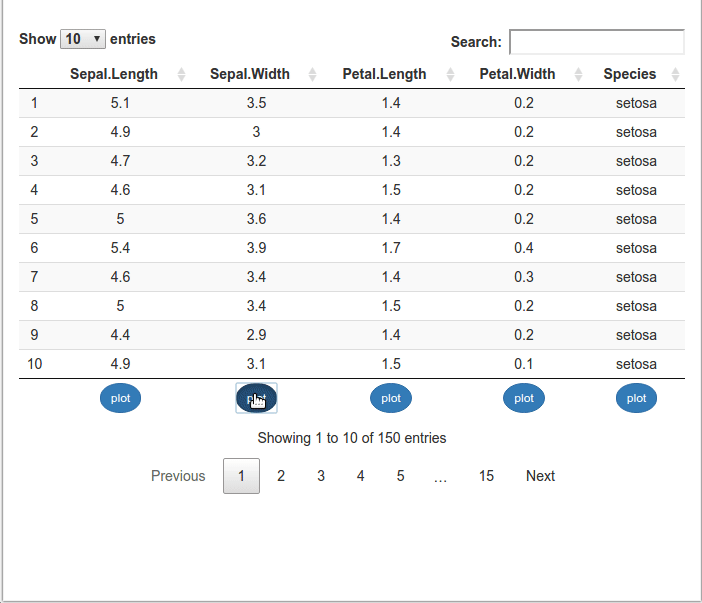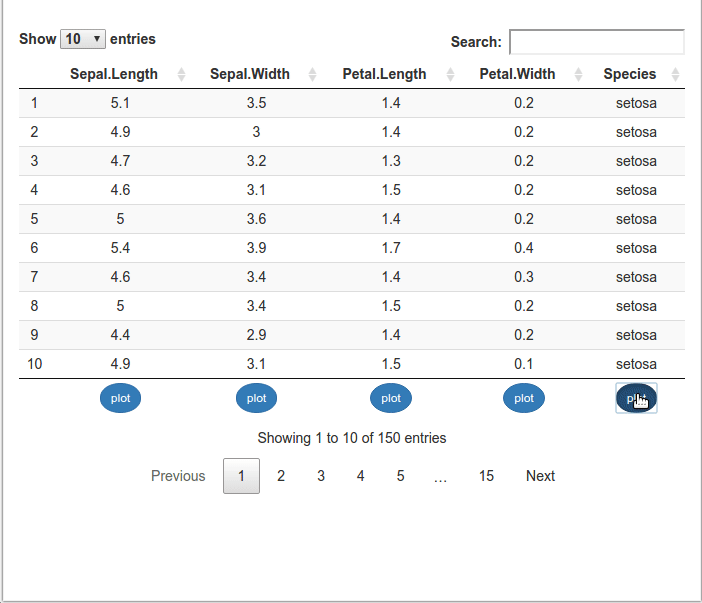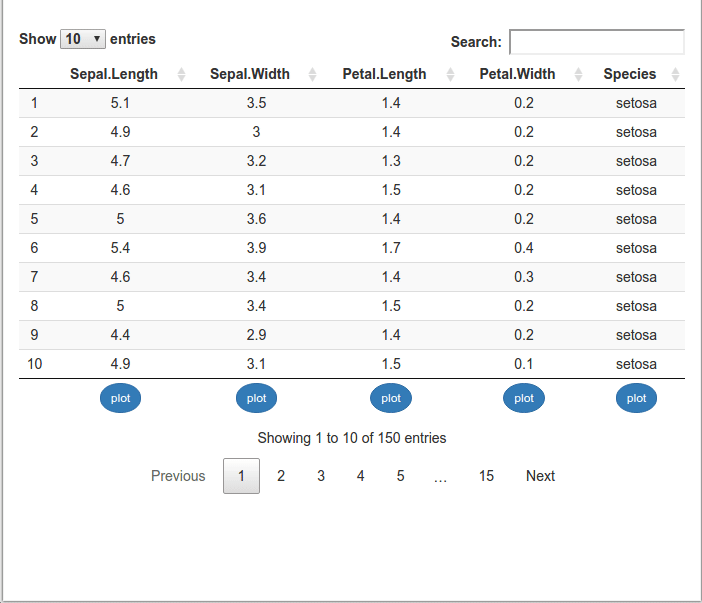
The following is the corresponding code:
library(shiny)
library(shinyBS)
library(DT)
library(ggplot2)
ui <- fluidPage(
uiOutput("modals"),
DTOutput("table")
)
server <- function(input, output, session){
dat <- iris
buttons <- lapply(1:ncol(dat), function(i){
actionButton(
paste0("this_id_is_not_used",i),
"plot",
class = "btn-primary btn-sm",
style = "border-radius: 50%;",
onclick = sprintf(
"Shiny.setInputValue('button', %d, {priority:'event'});
$('#modal%d').modal('show');", i, i)
)
})
output[["table"]] <- renderDT({
sketch <- tags$table(
class = "row-border stripe hover compact",
tableHeader(c("", names(dat))),
tableFooter(c("", buttons))
)
datatable(
dat, container = sketch,
options =
list(
columnDefs = list(
list(
className = "dt-center",
targets = "_all"
)
)
)
)
})
# modals ####
output[["modals"]] <- renderUI({
lapply(1:ncol(dat), function(i){
bsModal(
id = paste0("modal",i),
title = names(dat)[i],
trigger = paste0("this_is_not_used",i),
if(is.numeric(dat[[i]]) && length(unique(dat[[i]]))>19){
fluidRow(
column(5, radioButtons(paste0("radio",i), "",
c("density", "histogram"), inline = TRUE)),
column(7,
conditionalPanel(
condition = sprintf("input.radio%d=='histogram'",i),
sliderInput(paste0("slider",i), "Number of bins",
min = 5, max = 100, value = 30)
))
)
},
plotOutput(paste0("plot",i))
)
})
})
# plots in modals ####
for(i in 1:ncol(dat)){
local({
ii <- i
output[[paste0("plot",ii)]] <- renderPlot({
if(is.numeric(dat[[ii]]) && length(unique(dat[[ii]]))>19){
if(input[[paste0("radio",ii)]] == "density"){
ggplot(dat, aes_string(names(dat)[ii])) +
geom_density(fill = "seashell", color = "seashell") +
stat_density(geom = "line", size = 1) +
theme_bw() + theme(axis.title = element_text(size = 16))
}else{
ggplot(dat, aes_string(names(dat)[ii])) +
geom_histogram(bins = input[[paste0("slider",ii)]]) +
theme_bw() + theme(axis.title = element_text(size = 16))
}
}else{
dat[[".x"]] <-
factor(dat[[ii]], levels = names(sort(table(dat[[ii]]),
decreasing=TRUE)))
gg <- ggplot(dat, aes(.x)) + geom_bar() +
geom_text(stat="count", aes(label=..count..), vjust=-0.5) +
xlab(names(dat)[ii]) + theme_bw()
if(max(nchar(levels(dat$.x)))*nlevels(dat$.x)>40){
gg <- gg + theme(axis.text.x =
element_text(size = 12, angle = 45,
vjust = 0.5, hjust = 0.5))
}else{
gg <- gg + theme(axis.text.x = element_text(size = 12))
}
gg + theme(axis.title = element_text(size = 16))
}
})
})
}
}
shinyApp(ui, server)
My goal is to make an rshiny to replace the iris data above with the data uploaded by the user himself. Here, I use rshiny's fileinput. The following is the code
ui <- fluidPage(
fileInput("upload", NULL, accept = c(".csv", ".tsv")),
)
server <- function(input, output, session) {
data <- reactive({
req(input$upload)
ext <- tools::file_ext(input$upload$name)
switch(ext,
csv = vroom::vroom(input$upload$datapath, delim = ","),
tsv = vroom::vroom(input$upload$datapath, delim = "\t"),
validate("Invalid file; Please upload a .csv or .tsv file")
)
})
}
shinyApp(ui, server)
I have tried two methods, one is to name the data uploaded by the user DAT and replace it directly, and the other is to change the following dat to dat () after the user uploads the data, but all of them report errors. If anyone can help me solve this problem, I will be grateful