Here is a sample of my dataframe. Named "sampleforR"
data.frame(
patientno = c(1,2,3,4,5,6,7,8,9,10,11,12,13,
14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,
30,31,32,33,34,35,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,
NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,
NA,NA,NA),
time = c(315,184,567,444,465,1339,541,259,
644,547,773,475,242,699,539,1283,511,552,1066,178,114,
316,1116,627,518,380,395,146,410,132,249,30,168,
235,263,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,
NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA),
status = c(2,2,1,2,2,2,1,2,2,2,2,2,2,2,1,
2,1,2,2,2,2,1,2,2,2,2,2,1,2,2,2,2,1,1,1,2,
2,1,2,2,2,2,2,2,2,2,1,1,2,1,2,2,2,2,1,2,2,
2,2,2,1,2,1)
)
now, when I enter the following code I get the table below, I cannot remove the text 'STRATA' & 'ALL' on the at risk table. How can I make it go away?? it's spoiling the aesthetics. Thanks
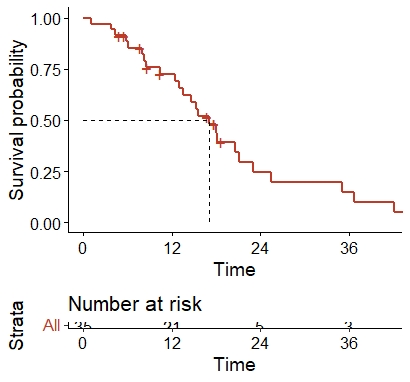
Thanks.
ggsurvplot(survfit(Surv(time,status) ~1, data = sampleforR), palette = "nejm", surv.median.line = "hv", xscale = "d_m", legend = "none", break.x.by = 365.25, conf.int=FALSE, risk.table = T)
