Hi!
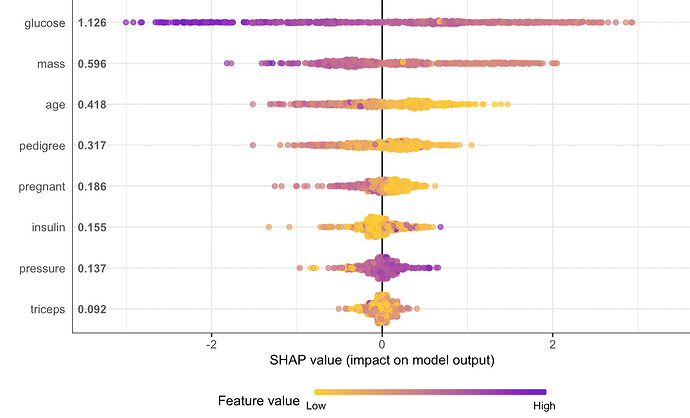
My shap values seems to be backwards when using xgboost classification in tidymodels. The results implies that a high blood glucose is correlated with lower diabetes risk. I can't make sense of it. Using other frameworks (ex standard xgboost-package) the shap values are logical, but not when using tidymodels. I suppose there is some issue with how I extract the engine or the training data when prepping for shap?
library(tidyverse)
library(tidymodels)
library(SHAPforxgboost)
library(mlbench)
#Loading data
data(PimaIndiansDiabetes)
data <- PimaIndiansDiabetes
#Converting target variable to factor
data$diabetes <- ifelse(data$diabetes == "pos", 1, 0)
data$diabetes <- as.factor(data$diabetes)
#Train/test split
set.seed(1992)
dm_split <- initial_split(data, strata = diabetes, prop = 0.8)
dm_train <- training(dm_split)
dm_test <- testing(dm_split)
#Recipe
dm_rec <- recipe(diabetes ~., data = dm_train) %>%
step_zv(all_numeric())
dm_prep <- prep(dm_rec)
#Model specification
dm_spec <- boost_tree()%>%
set_engine("xgboost") %>%
set_mode("classification")
#Workflow
dm_wf <- workflow(
dm_rec,
dm_spec
)
#CV-folds
set.seed(1992)
dm_folds <- vfold_cv(data = dm_train, strata = diabetes, v = 5)
dm_res <-
dm_wf %>%
fit_resamples(
resamples = dm_folds,
metrics = metric_set(
recall, precision, f_meas,
accuracy, kap,
roc_auc, sens, spec),
control = control_resamples(save_pred = TRUE)
)
dm_res %>% collect_metrics(summarize = TRUE)
best_auc <- select_best(dm_res, "roc_auc")
#Finalize workflow
dm_fit <- dm_wf %>%
finalize_workflow(best_auc) %>%
last_fit(dm_split)
#Preparing data for shap
dm_shap <-
shap.prep(
xgb_model = extract_fit_engine(dm_fit),
X_train = bake(dm_prep,
has_role("predictor"),
new_data = NULL,
composition = "matrix"
))
#Shap summary
shap.plot.summary(dm_shap)