Hello Andres, I'll do my best:
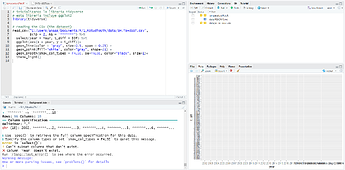
My code:
library(tidyverse)
# reading the CSV
read_csv("C:/Users/anns/Documents/R/UTN/data/SH.Ts+dSST.csv",
skip = 1, na = "*******") +
select(year = Year, t_diff = DJF) %>%
ggplot(aes(x = year, y = t_diff)) +
geom_line(color = "gray", size=0.5, span = 0.25) +
geom_point(fill="white", color="gray", shape=21) +
geom_smooth(show_col_types = FALSE, se=FALSE, color="black", size=1) +
theme_light()
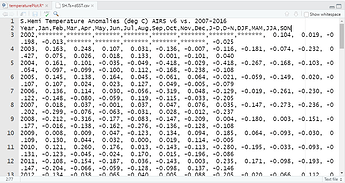
The dataset:
S.Hemi Temperature Anomalies (deg C) AIRS v6 vs. 2007-2016
Year,Jan,Feb,Mar,Apr,May,Jun,Jul,Aug,Sep,Oct,Nov,Dec,J-D,D-N,DJF,MAM,JJA,SON
2002,*******,*******,*******,*******,*******,*******,*******,*******, 0.104, 0.019, -0.198, -0.013,*******,*******,*******,*******,*******, -0.025
2003, 0.163, 0.248, 0.107, 0.031, -0.136, -0.007, -0.116, -0.181, -0.074, -0.232, 0.427, 0.075, 0.026, 0.018, 0.133, 0.001, -0.101, 0.040
2004, 0.161, 0.101, -0.035, -0.049, -0.418, -0.029, -0.418, -0.267, -0.168, -0.103, -0.054, 0.097, -0.099, -0.100, 0.112, -0.168, -0.238, -0.108
2005, 0.145, 0.138, 0.164, 0.045, -0.061, 0.064, -0.021, -0.059, -0.149, 0.020, -0.107, 0.107, 0.024, 0.023, 0.127, 0.049, -0.005, -0.079
2006, 0.136, 0.114, 0.030, -0.056, -0.319, 0.048, -0.129, -0.019, -0.261, -0.230, -0.122, -0.148, -0.080, -0.059, 0.119, -0.115, -0.033, -0.205
2007, 0.018, 0.037, -0.001, 0.037, 0.047, 0.076, 0.035, -0.147, -0.273, -0.236, -0.202, -0.299, -0.076, -0.063, -0.031, 0.028, -0.012, -0.237
2008, -0.212, -0.316, -0.177, -0.083, -0.147, -0.209, 0.004, -0.180, 0.003, -0.151, -0.177, -0.016, -0.138, -0.162, -0.276, -0.136, -0.128, -0.108
2009, 0.008, 0.009, 0.047, -0.123, 0.134, 0.094, 0.185, 0.064, -0.093, -0.030, 0.109, 0.130, 0.044, 0.032, 0.000, 0.019, 0.114, -0.005
2010, 0.121, 0.260, 0.176, 0.013, -0.143, -0.113, -0.280, -0.195, -0.033, -0.093, -0.131, -0.123, -0.045, -0.024, 0.170, 0.015, -0.196, -0.086
2011, -0.108, -0.154, -0.187, 0.036, -0.143, 0.003, 0.235, 0.171, -0.098, -0.193, -0.147, -0.204, -0.066, -0.059, -0.128, -0.098, 0.137, -0.146
2012, -0.134, -0.038, -0.065, -0.040, 0.005, -0.088, -0.205, -0.020, -0.066, 0.112, 0.071, 0.046, -0.035, -0.056, -0.125, -0.034, -0.105, 0.039
2013, 0.113, 0.043, -0.015, -0.012, -0.118, 0.117, -0.129, 0.027, 0.240, 0.026, 0.023, 0.075, 0.033, 0.030, 0.068, -0.048, 0.005, 0.097
2014, 0.001, -0.161, -0.058, 0.003, 0.148, -0.043, -0.076, -0.026, 0.198, 0.110, 0.071, 0.043, 0.017, 0.020, -0.028, 0.031, -0.049, 0.126
2015, 0.047, 0.112, 0.028, -0.116, -0.063, 0.007, -0.068, 0.016, 0.022, 0.169, 0.082, 0.218, 0.038, 0.023, 0.068, -0.050, -0.015, 0.091
2016, 0.147, 0.208, 0.252, 0.286, 0.280, 0.156, 0.300, 0.291, 0.099, 0.287, 0.301, 0.129, 0.228, 0.235, 0.191, 0.273, 0.249, 0.229
2017, 0.161, 0.255, 0.258, 0.134, 0.306, 0.170, 0.217, 0.106, 0.087, 0.236, 0.046, -0.013, 0.164, 0.175, 0.182, 0.233, 0.164, 0.123
2018, 0.101, 0.010, 0.118, 0.278, 0.059, 0.166, 0.099, 0.062, -0.009, 0.085, 0.065, 0.204, 0.103, 0.085, 0.033, 0.152, 0.109, 0.047
2019, 0.209, 0.281, 0.285, 0.292, 0.222, 0.302, 0.406, 0.233, 0.188, 0.340, 0.315, 0.378, 0.288, 0.273, 0.231, 0.266, 0.314, 0.281
2020, 0.250, 0.060, 0.047, 0.288, 0.198, 0.275, 0.288, 0.018, 0.188, -0.030, 0.114, -0.121, 0.131, 0.173, 0.229, 0.177, 0.193, 0.091
2021, -0.059, -0.129, -0.030, -0.001, -0.005, 0.020, 0.200, 0.072, 0.106, -0.210, -0.195, 0.058, -0.014, -0.029, -0.103, -0.012, 0.097, -0.100
2022, 0.063, 0.032, 0.137, -0.180, -0.057, 0.097, 0.004, -0.204,*******,*******,*******,*******,*******,*******, 0.051, -0.033, -0.034,*******
S.Hemi Temperature Anomalies (deg C) AIRS v7 vs. 2007-2016
Year,Jan,Feb,Mar,Apr,May,Jun,Jul,Aug,Sep,Oct,Nov,Dec,J-D,D-N,DJF,MAM,JJA,SON
2002,*******,*******,*******,*******,*******,*******,*******,*******, 0.316, 0.219, 0.080, 0.170,*******,*******,*******,*******,*******, 0.205
2003, 0.234, 0.269, 0.197, 0.244, 0.131, 0.189, 0.080, -0.011, 0.087, -0.075, 0.606, 0.247, 0.183, 0.177, 0.224, 0.191, 0.086, 0.206
2004, 0.269, 0.145, 0.061, 0.123, -0.287, 0.118, -0.353, -0.214, -0.011, 0.047, 0.082, 0.190, 0.014, 0.019, 0.220, -0.034, -0.150, 0.039
2005, 0.234, 0.208, 0.221, 0.181, 0.071, 0.162, 0.044, 0.035, -0.067, 0.108, -0.066, 0.126, 0.105, 0.110, 0.210, 0.158, 0.080, -0.008
2006, 0.151, 0.118, 0.063, 0.006, -0.219, 0.092, -0.073, 0.025, -0.208, -0.120, -0.043, -0.103, -0.026, -0.007, 0.132, -0.050, 0.015, -0.124
2007, 0.036, 0.049, -0.018, 0.079, 0.126, 0.131, 0.064, -0.115, -0.174, -0.200, -0.184, -0.296, -0.042, -0.026, -0.006, 0.062, 0.027, -0.186
2008, -0.224, -0.325, -0.195, -0.064, -0.141, -0.223, 0.086, -0.136, -0.007, -0.141, -0.148, -0.033, -0.129, -0.151, -0.282, -0.134, -0.091, -0.099
2009, 0.023, -0.033, 0.289, -0.080, 0.134, 0.128, 0.255, 0.073, -0.090, -0.025, 0.133, 0.084, 0.074, 0.065, -0.014, 0.114, 0.152, 0.006
2010, 0.121, 0.218, 0.145, 0.033, -0.082, -0.061, -0.277, -0.171, -0.033, -0.109, -0.145, -0.116, -0.040, -0.023, 0.141, 0.032, -0.170, -0.096
2011, -0.065, -0.125, -0.218, 0.036, -0.109, 0.028, 0.243, 0.151, -0.078, -0.165, -0.147, -0.217, -0.056, -0.047, -0.102, -0.097, 0.141, -0.130
2012, -0.221, -0.055, -0.093, -0.038, 0.032, -0.045, -0.204, -0.007, -0.099, 0.104, -0.001, 0.026, -0.050, -0.070, -0.164, -0.033, -0.086, 0.001
2013, 0.083, 0.024, -0.040, 0.008, -0.128, 0.144, -0.169, 0.005, 0.236, -0.016, -0.019, 0.044, 0.014, 0.013, 0.045, -0.053, -0.007, 0.067
2014, -0.001, -0.185, -0.065, 0.003, 0.144, -0.009, -0.071, 0.034, 0.201, 0.125, 0.061, 0.032, 0.023, 0.024, -0.047, 0.027, -0.015, 0.129
2015, 0.042, 0.154, 0.003, -0.156, -0.108, -0.101, -0.140, -0.044, -0.008, 0.175, 0.157, 0.291, 0.022, 0.001, 0.076, -0.087, -0.095, 0.108
2016, 0.205, 0.277, 0.191, 0.180, 0.132, 0.008, 0.213, 0.211, 0.053, 0.251, 0.294, 0.183, 0.183, 0.192, 0.258, 0.167, 0.144, 0.199
2017, 0.224, 0.267, 0.227, 0.066, 0.218, -0.005, 0.108, 0.017, 0.071, 0.142, -0.001, -0.010, 0.110, 0.127, 0.225, 0.170, 0.040, 0.071
2018, 0.112, 0.017, 0.067, 0.205, 0.021, 0.049, -0.003, -0.040, -0.064, 0.021, 0.037, 0.213, 0.053, 0.034, 0.040, 0.098, 0.002, -0.002
2019, 0.235, 0.341, 0.272, 0.190, 0.051, 0.072, 0.193, 0.149, 0.075, 0.249, 0.300, 0.348, 0.206, 0.195, 0.263, 0.171, 0.138, 0.208
2020, 0.249, 0.126, 0.050, 0.225, 0.110, 0.163, 0.148, -0.066, 0.174, 0.002, 0.159, -0.141, 0.100, 0.141, 0.241, 0.128, 0.082, 0.112
2021, -0.104, -0.141, -0.060, -0.060, -0.069, -0.097, 0.148, 0.079, 0.178, 0.018, -0.017, 0.228, 0.009, -0.022, -0.128, -0.063, 0.043, 0.060
2022, 0.226, 0.234, 0.241, -0.244, 0.096, 0.173, 0.124, -0.100,*******,*******,*******,*******,*******,*******, 0.229, 0.031, 0.066,*******
S.Hemi Temperature Anomalies (deg C) GHCNv4/ERSSTv5 vs. 2007-2016
Year,Jan,Feb,Mar,Apr,May,Jun,Jul,Aug,Sep,Oct,Nov,Dec,J-D,D-N,DJF,MAM,JJA,SON
2002, 0.006, -0.052, 0.127, 0.166, 0.190, -0.134, 0.022, -0.098, 0.024, -0.032, -0.163, -0.057, -0.000, 0.000, -0.033, 0.161, -0.070, -0.057
2003, 0.055, 0.125, 0.007, 0.004, -0.081, -0.095, -0.061, -0.181, -0.148, -0.110, -0.153, -0.000, -0.053, -0.058, 0.041, -0.023, -0.112, -0.137
2004, -0.016, 0.048, -0.122, 0.030, -0.364, -0.104, -0.510, -0.246, -0.203, -0.068, -0.085, 0.056, -0.132, -0.137, 0.010, -0.152, -0.287, -0.119
2005, 0.029, 0.005, 0.018, -0.078, -0.095, -0.008, -0.059, -0.095, -0.092, -0.026, -0.145, 0.011, -0.045, -0.041, 0.030, -0.052, -0.054, -0.088
2006, -0.044, 0.023, -0.044, -0.149, -0.390, -0.009, -0.159, 0.124, -0.135, -0.146, -0.025, -0.090, -0.087, -0.079, -0.003, -0.194, -0.015, -0.102
2007, 0.066, 0.023, -0.026, -0.045, 0.070, 0.061, 0.003, -0.145, -0.085, -0.236, -0.199, -0.290, -0.067, -0.050, -0.000, -0.000, -0.027, -0.173
2008, -0.243, -0.310, -0.178, -0.068, -0.201, -0.152, 0.056, -0.286, 0.036, -0.099, -0.143, -0.088, -0.140, -0.156, -0.281, -0.149, -0.127, -0.068
2009, -0.049, -0.121, -0.039, 0.027, 0.056, 0.104, 0.255, 0.032, -0.019, -0.071, 0.191, 0.174, 0.045, 0.023, -0.086, 0.015, 0.130, 0.033
2010, 0.178, 0.253, 0.223, 0.069, -0.012, 0.006, -0.172, -0.139, -0.058, -0.066, -0.134, -0.117, 0.003, 0.027, 0.202, 0.094, -0.102, -0.086
2011, -0.085, -0.083, -0.099, -0.037, -0.179, -0.017, 0.147, 0.168, -0.221, -0.172, -0.048, -0.196, -0.068, -0.062, -0.095, -0.105, 0.100, -0.147
2012, -0.219, -0.082, -0.081, -0.114, 0.008, -0.159, -0.223, -0.069, -0.043, 0.096, 0.038, 0.094, -0.063, -0.087, -0.166, -0.062, -0.150, 0.031
2013, 0.050, -0.020, 0.011, -0.092, -0.149, 0.102, -0.034, 0.034, 0.226, -0.026, -0.009, 0.038, 0.011, 0.016, 0.041, -0.076, 0.034, 0.064
2014, 0.023, -0.099, -0.160, 0.019, 0.237, 0.020, -0.142, 0.133, 0.298, 0.103, -0.031, -0.050, 0.029, 0.037, -0.012, 0.032, 0.004, 0.123
2015, 0.006, 0.132, 0.061, 0.004, -0.032, 0.060, 0.004, -0.024, -0.096, 0.313, 0.179, 0.267, 0.073, 0.046, 0.029, 0.011, 0.013, 0.132
2016, 0.272, 0.307, 0.289, 0.236, 0.200, -0.027, 0.106, 0.297, -0.037, 0.157, 0.156, 0.169, 0.177, 0.185, 0.282, 0.242, 0.125, 0.092
2017, 0.137, 0.268, 0.280, 0.196, 0.314, 0.002, 0.128, 0.112, -0.082, 0.170, -0.018, -0.043, 0.122, 0.140, 0.192, 0.263, 0.081, 0.023
2018, -0.029, -0.036, 0.080, 0.323, 0.052, 0.180, 0.210, 0.051, 0.018, 0.216, 0.121, 0.220, 0.117, 0.095, -0.036, 0.152, 0.147, 0.118
2019, 0.145, 0.290, 0.272, 0.289, 0.134, 0.113, 0.318, 0.194, 0.021, 0.138, 0.229, 0.260, 0.200, 0.197, 0.218, 0.231, 0.208, 0.129
2020, 0.227, 0.295, 0.156, 0.366, 0.171, 0.198, 0.160, 0.008, 0.155, -0.026, 0.039, -0.125, 0.135, 0.167, 0.261, 0.231, 0.122, 0.056
2021, -0.159, -0.161, 0.043, -0.112, -0.056, -0.036, 0.229, -0.009, 0.172, 0.098, 0.001, 0.047, 0.005, -0.009, -0.148, -0.042, 0.062, 0.090
2022, 0.045, 0.127, 0.143, 0.085, 0.078, 0.208, 0.275, 0.110,*******,*******,*******,*******,*******,*******, 0.073, 0.102, 0.198,*******
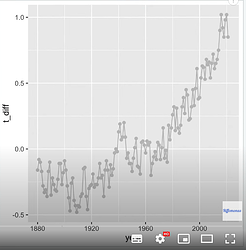
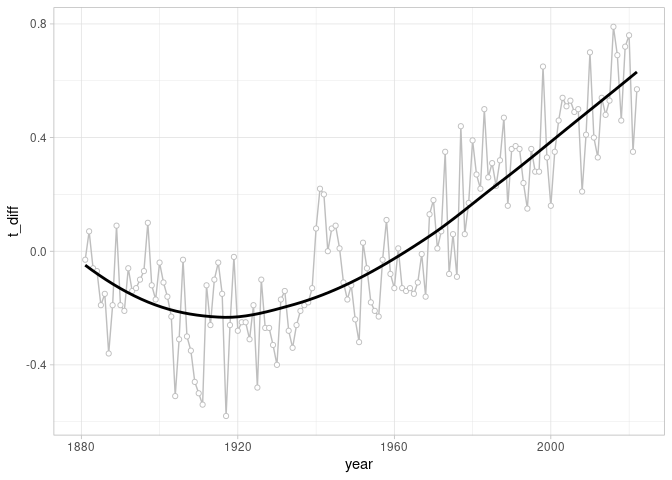
The outcome of my code: