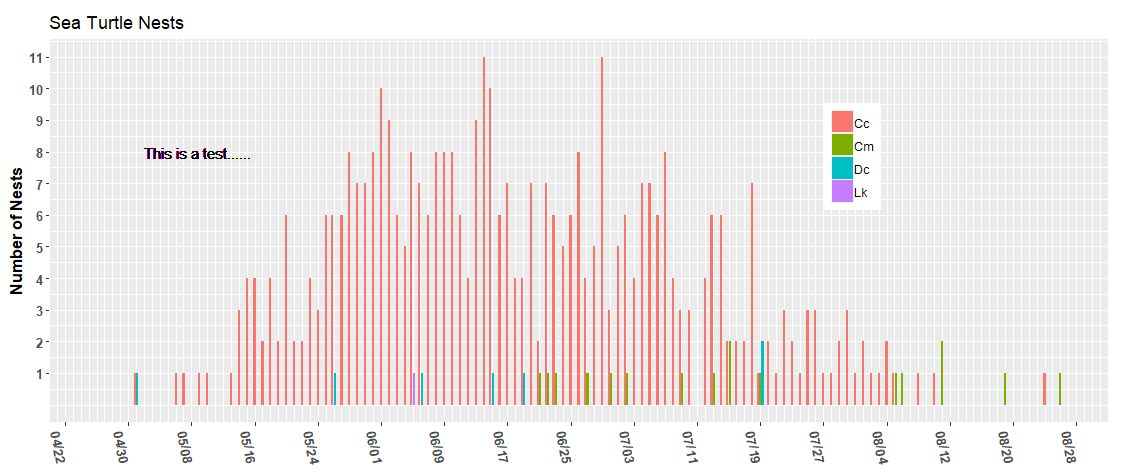
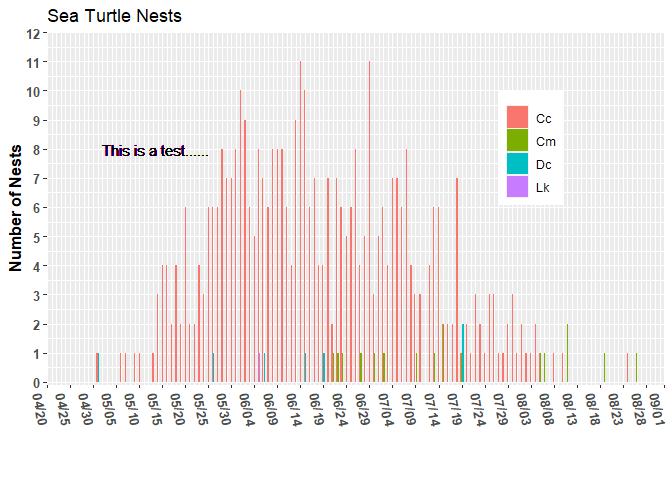
I thought I was getting a decent handle on working with dates. I have set start and end dates as 2022-04-20 and 2022-09-01, respectively. However, the chart appears to have them as 2022-04-13 and 2022-09-08. Not sure why. The text and legend positioning are off a bit too. However, I thought I would deal with that in a separate post once I have the limits working as desired.
library(tidyverse)
library("lubridate")
#> Loading required package: timechange
#>
#> Attaching package: 'lubridate'
#> The following objects are masked from 'package:base':
#>
#> date, intersect, setdiff, union
library("reprex")
start_date <- ymd("2022-04-20")
end_date <- ymd("2022-09-01")
position_date <- ymd("2022-07-27") #place on x-axis
date_length <-days(start_date %--% end_date )
position_date_len <- days(start_date %--% position_date)
relative_date <- position_date_len/date_length
start_date
#> [1] "2022-04-20"
end_date
#> [1] "2022-09-01"
position_date
#> [1] "2022-07-27"
date_length
#> [1] "11577600d 0H 0M 0S"
position_date_len
#> [1] "8467200d 0H 0M 0S"
relative_date
#> [1] 0.7313433
turtle_activity_gtm <- read_csv("https://www.dropbox.com/s/ujdyzqc74152325/turtle_activity_report.csv?dl=1")
#> Rows: 925 Columns: 62
#> ── Column specification ────────────────────────────────────────────────────────
#> Delimiter: ","
#> chr (22): beach, county, activity, ref_no, activity_comments, encountered?,...
#> dbl (31): uid, activity_no, nest_no, year, month, week, dayofyear, julianda...
#> lgl (6): final_treatment, light_management, relocation_reason, lost_nest, ...
#> date (3): activity_date, emerge_date, inventory_date
#>
#> ℹ Use `spec()` to retrieve the full column specification for this data.
#> ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
ggplot(data = (turtle_activity_gtm |> filter(activity=="N"))) +
geom_bar(aes(x = activity_date, fill = species),
position = position_dodge2(preserve = "single")) +
scale_y_continuous(breaks = 1:12) +
scale_x_date(date_breaks = "8 day",
date_labels = "%m/%d",
date_minor_breaks = "1 day",
limits = c( start_date, end_date)) +
labs(x="",y="Number of Nests", title="Sea Turtle Nests", caption=' ') +
theme(axis.text.y = element_text(face = "bold",
size = 10, angle = 0),
axis.text.x = element_text(face = "bold",
size = 10, angle = -80),
legend.position=c(x=relative_date, y=10/12),
legend.justification = c(0, 1), #upper left
legend.title = element_blank(),
legend.spacing.x = unit(0, "mm"),
legend.spacing.y = unit(0, "mm"),
axis.title = element_text(size = 12, face = "bold"),
plot.caption = element_text(size = 15)) +
geom_text(x=ymd("2022-05-02"), y=8, hjust = "left",label="This is a test......")
#> Warning: Removed 1 rows containing non-finite values (`stat_count()`).
Created on 2023-01-11 with reprex v2.0.2
Thanks,
Jeff