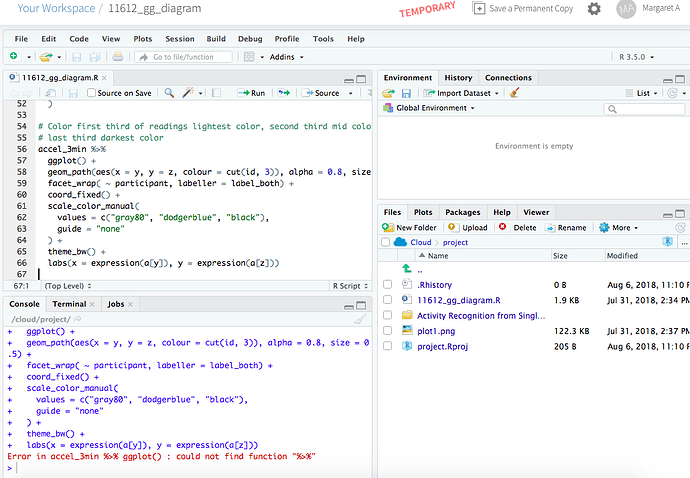
Yes, all the packages were successfully installed. I was using the script provided by you, i.e. the below:-
library(tidyverse)
library(ggplot2)
library(reprex)
#
# Example data file from a X2-2 accelerometer
file_loc <- 'http://www.gcdataconcepts.com/DATA-100.CSV'
#
# Function to load
load_accel <- function(fn, label = NA){
# Read header lines from the file
fc <- file(fn)
hh <- readLines(fc, n = 9)
close(fc)
# edit and split the header lines lines
hhl <- gsub(", ", ",", hh) %>% str_split(",")
# extract the gain amnd header names
gain <- hhl[[grep(";Gain", hhl)]][2]
heads <- hhl[[grep(";Headers", hhl)]][2:5]
#
# Read full data from the file:
atbl <- read_csv(fn, col_names = heads, comment = ";")
#
if( gain == "low" ){ conv_fac <- 1/6554 } else { conv_fac <- 1/13108 }
atbl %>% mutate( Axg = Ax * conv_fac,
Ayg = Ay * conv_fac,
Azg = Az * conv_fac, expt =label ) %>%
# drops expt if all NA, assumes other columns contain values
select_if(function(x){!all(is.na(x))})
}
#
# As you have two experiments/treatments, at 1m and 3m, I've plotted
# the same file twice, once for each. You would have two different file
# locations here.
#
# Assuming you have accelerometer data files at 1m and 3m, this
# code is what you need (with the file names modified to yours):
# acc_1m_and_3m <- bind_rows( load_accel( "my_1m_accel_data.csv", "1m" ),
# load_accel( "my_3m_accel_data.csv", "3m" ) )
#
# Because I am re-plotting the same data file twice in this example, I'm
# going to scale the 1m height data to make it less variable:
#
d_1m <- load_accel( file_loc, "1m" ) %>%
mutate( Axg = Axg*0.5,
Ayg = Ayg*0.5,
Azg = Azg*0.5 )
#> Parsed with column specification:
#> cols(
#> time = col_double(),
#> Ax = col_integer(),
#> Ay = col_integer(),
#> Az = col_integer()
#> )
# and then combine it with the 3m height data:
d_3m <- load_accel( file_loc, "3m" )
#> Parsed with column specification:
#> cols(
#> time = col_double(),
#> Ax = col_integer(),
#> Ay = col_integer(),
#> Az = col_integer()
#> )
acc_1m_and_3m <- bind_rows( d_1m, d_3m )
#
# Now we're ready to plot the data in acc_1m_and_3m
#
ggplot(acc_1m_and_3m) +
geom_path(aes(x = Azg, y = Ayg,
# I'm generating a factor from expt with the levels in
# a specific order so that the more variable 3m data will
# be plotted beneath the 1m data, as in the examples
colour = factor(expt,levels=c("3m","1m"))), size = 0.5) +
coord_fixed() + theme_bw() +
scale_x_continuous(name = expression(A[z] (g))) +
scale_y_continuous(name = expression(A[y] (g))) +
scale_colour_manual(name = "Height", values=c("dodgerblue","black"))
I am still trying to understand what reprex and how reprex works, it will take me some times to work on it. I will update my reprex outcome here later. Thank you so much.