Hi Jonspring,
I have some doubts, please correct me, if I m wrong.
sensor_data <- data.table::fread("org_data.csv") %>%
# convert localminute to datetime (fread imports it as character)
mutate(localminute = lubridate::as_datetime(localminute)) %>%
group_by(dataid) %>%
arrange(localminute, .by_group = TRUE) %>%
mutate(interval_hr = (localminute - lag(localminute)) / lubridate::dhours(1),
meter_change = meter_value - lag(meter_value)) %>% ungroup()
print(sensor_data)
# A tibble: 1,584,823 x 5
localminute dataid meter_value interval_hr meter_change
<dttm> <int> <int> <dbl> <int>
1 2015-10-01 05:14:44 35 93470 NA NA
2 2015-10-01 05:42:34 35 93470 0.464 0
3 2015-10-01 07:02:37 35 93470 1.33 0
4 2015-10-01 07:12:38 35 93470 0.167 0
5 2015-10-01 07:20:36 35 93470 0.133 0
6 2015-10-01 07:23:39 35 93470 0.0508 0
7 2015-10-01 08:59:41 35 93470 1.60 0
8 2015-10-01 09:30:40 35 93470 0.516 0
9 2015-10-01 09:34:37 35 93470 0.0658 0
10 2015-10-01 10:14:35 35 93470 0.666 0
# ... with 1,584,813 more rows
"","localminute","dataid","meter_value","interval_hr","meter_change" # to see 1,584,813 more rows, i export to csv file
"12",2015-10-01 11:23:36,35,93472,1.10111111111111,2
```````````````````````````````````````````````
What is the meaning of interval_hr = (localminute - lag(localminute)) / lubridate::dhours(1)?
How the meter_change is calculated, as I saw some dataid has value in meter_change?
Ungroup() meaning?
'function: sensor_data' is listing all the meter_value with meter_changes, means that: including spiky data and non-spiky data?
the list in 'function: noisy_readings' is filtered from 'sensor_data function'?
# Which have noisy data? (assuming a negative change indicates an error)
noisy_readings <-
sensor_data %>%
filter(meter_change < -100) # Setting here to ignore small changes
> print(noisy_readings)
# A tibble: 1,094 x 5
localminute dataid meter_value interval_hr meter_change
<dttm> <int> <int> <dbl> <int>
1 2015-12-07 23:19:59 1185 144548 0.231 -17798 #this data was no in "sensor_data"function, when i extract
2 2015-12-08 00:21:50 1185 144550 0.0928 -17798
3 2015-12-08 01:58:12 1185 144558 0.0978 -17792
4 2015-12-08 02:12:12 1185 144558 0.0661 -17792
5 2015-12-08 02:34:59 1185 144558 0.0292 -17792
6 2015-12-08 03:20:06 1185 144558 0.699 -17792
7 2015-12-08 03:58:58 1185 144558 0.364 -17792
8 2015-12-08 05:09:55 1185 144558 1.06 -17792
9 2015-12-08 07:05:02 1185 144560 0.0325 -17792
10 2015-12-08 07:52:51 1185 144592 0.0453 -17760
# ... with 1,084 more rows
> noisy_meters <- unique(noisy_readings$dataid)
> print(noisy_meters)
[1] 1185 1556 2335 2449 3134 3544 4514 5129 5403 6836 7030 7117 8156 9134 9639 9982
'function: noisy_meters' is listing the noisy dataid from the noisy_readings?
Is that what unique function meaning?
> noisy_ranges <-
noisy_readings %>%
group_by(dataid) %>%
summarize(min_range = min(localminute) - 60*60*24*3,
max_range = max(localminute) + 60*60*24*3)
'function: noisy_ranges' meaning arranging a list of noisy dataid, starting with their minimum localminute to maximum localminute?
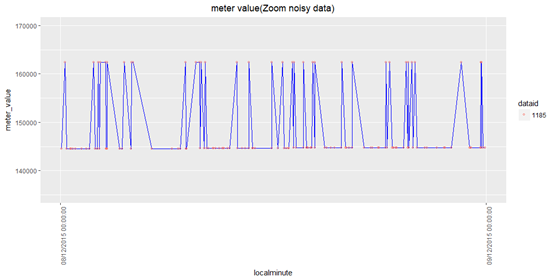
eg. for dataid =1185, arrange its' noisy data start from miniimum localminute (2015-12-04 23:19:59) to maximum localminute (2015-12-16 07:27:54)?
As u said "606024 is one day", but why multiple by 3?
> print(noisy_ranges)
> # A tibble: 16 x 3
> dataid min_range max_range
> <int> <dttm> <dttm>
> 1 1185 2015-12-04 23:19:59 2015-12-16 07:27:54
> 2 1556 2015-12-05 08:36:42 2015-12-14 08:14:36
> 3 2335 2015-12-05 22:33:26 2015-12-15 05:52:27
> 4 2449 2015-12-04 23:07:19 2015-12-16 07:34:19
> 5 3134 2015-12-05 12:56:40 2015-12-15 23:10:28
> 6 3544 2015-12-05 01:49:52 2015-12-16 00:48:35
> 7 4514 2015-12-04 22:41:04 2015-12-16 07:19:59
> 8 5129 2015-12-04 23:10:13 2015-12-16 08:02:14
> 9 5403 2015-12-04 23:42:18 2015-12-16 06:38:00
> 10 6836 2015-12-05 02:07:34 2015-12-16 05:56:47
> 11 7030 2015-12-04 23:54:21 2015-12-16 07:20:11
> 12 7117 2015-12-04 23:15:09 2015-12-16 07:12:02
> 13 8156 2015-12-04 22:47:45 2015-12-16 06:53:41
> 14 9134 2015-12-04 23:16:58 2015-12-16 08:01:12
> 15 9639 2015-12-19 06:27:07 2015-12-25 17:54:52
> 16 9982 2015-12-06 02:31:41 2015-12-14 18:29:49
> noisy_context <-
sensor_data %>%
left_join(noisy_ranges) %>%
filter(localminute >= min_range,
localminute <= max_range)
> print(noisy_context)
# A tibble: 13,913 x 7
localminute dataid meter_value interval_hr meter_change min_range
<dttm> <int> <int> <dbl> <int> <dttm>
1 2015-12-04 23:22:06 1185 143862 0.266 0 2015-12-04 23:19:59
2 2015-12-04 23:36:07 1185 143864 0.234 2 2015-12-04 23:19:59
3 2015-12-04 23:51:06 1185 143864 0.250 0 2015-12-04 23:19:59
4 2015-12-05 00:19:02 1185 143868 0.466 4 2015-12-04 23:19:59
5 2015-12-05 00:42:54 1185 143874 0.398 6 2015-12-04 23:19:59
6 2015-12-05 00:45:58 1185 143874 0.0511 0 2015-12-04 23:19:59
7 2015-12-05 00:57:53 1185 143874 0.199 0 2015-12-04 23:19:59
8 2015-12-05 01:14:53 1185 143876 0.283 2 2015-12-04 23:19:59
9 2015-12-05 01:29:54 1185 143876 0.250 0 2015-12-04 23:19:59
10 2015-12-05 01:31:57 1185 143876 0.0342 0 2015-12-04 23:19:59
# ... with 13,903 more rows, and 1 more variable: max_range <dttm>
I am confused about this 'function:noisy_context'. Why we need to create this 'noisy_context' data frame?
I tried plotting only one noisy dataid, what is the meaning of meter_value <= lead(meter_value)?
Looks like the data are overlapping, so couldnot clearly see the 3 different colours
ggplot(noisy_context %>% filter(dataid %in% c(4514)), aes(localminute, meter_value,
group = dataid, label = dataid,
color = meter_value <= lead(meter_value))) +
geom_point(shape = 1, alpha = 0.4) +
geom_line(alpha = 0.3) +
scale_y_continuous(labels = scales::comma) +
scale_x_datetime(date_breaks = "1 day", date_labels = "%b\n%d") +
facet_wrap(~dataid, scales = "free")