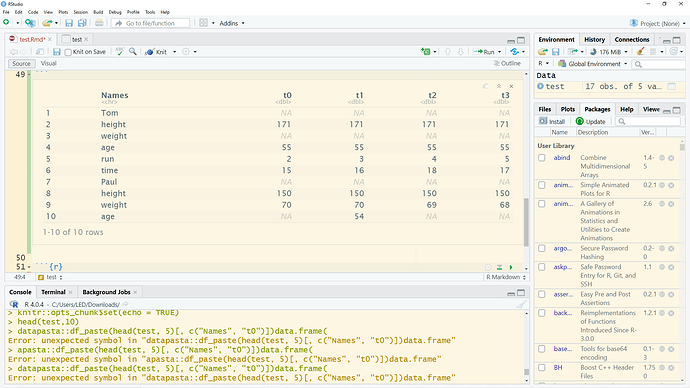
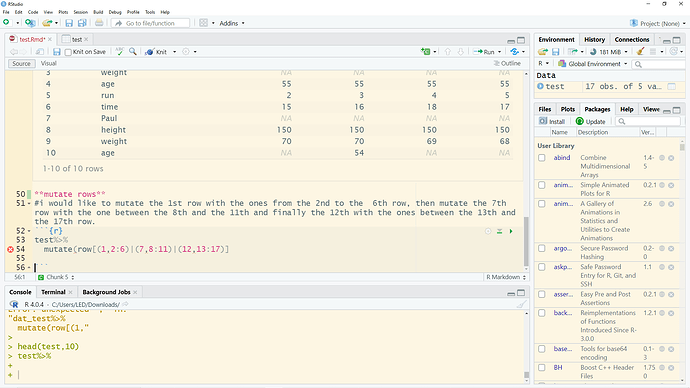
Hi FactOREO,
thanks! Let me try...
structure(list(Names = c("Tom", "height", "weight", "age", "run",
"time", "Paul", "height", "weight", "age", "time", "Steven",
"height", "weight", "age", "run", "time"), t0 = c(NA, 171, NA,
55, 2, 15, NA, 150, 70, NA, 15, NA, 171, 60, 55, 7, 15), t1 = c(NA,
171, NA, 55, 3, 16, NA, 150, 70, 54, 14, NA, NA, NA, NA, NA,
16), t2 = c(NA, 171, NA, 55, 4, 18, NA, 150, 69, NA, 13, NA,
171, 60, 55, 8, 17), t3 = c(NA, 171, NA, 55, 5, 17, NA, 150,
68, NA, NA, NA, 171, 60, NA, 8, 18)), class = c("tbl_df", "tbl",
"data.frame"), row.names = c(NA, -17L))
I hope it worked.
My wanted outcome for my example would be (see below), to create a new column with the names of the people.
structure(list(Names = c("Tom", "Tom", "Tom", "Tom", "Tom", "Paul",
"Paul", "Paul", "Paul", "Steven", "Steven", "Steven", "Steven",
"Steven"), variables = c("height", "weight", "age", "run", "time",
"height", "weight", "age", "time", "height", "weight", "age",
"run", "time"), t0 = c(171, NA, 55, 2, 15, 150, 70, NA, 15, 171,
60, 55, 7, 15), t1 = c(171, NA, 55, 3, 16, 150, 70, 54, 14, NA,
NA, NA, NA, 16), t2 = c(171, NA, 55, 4, 18, 150, 69, NA, 13,
171, 60, 55, 8, 17), t3 = c(171, NA, 55, 5, 17, 150, 68, NA,
NA, 171, 60, NA, 8, 18)), class = c("tbl_df", "tbl", "data.frame"
), row.names = c(NA, -14L))
Many thanks in advance