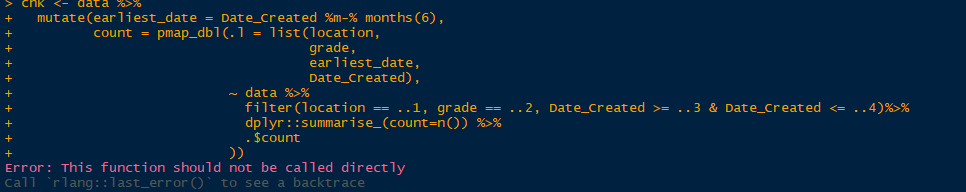
data$Date_Created = ymd(data$Date_Created)
chk <- data %>%
mutate(earliest_date = Date_Created %m-% months(6),
count = pmap_dbl(.l = list(location,
grade,
earliest_date,
Date_Created),
~ data %>%
filter(location == ..1, grade == ..2, Date_Created >= ..3 & Date_Created <= ..4)%>%
dplyr::summarise_(count=n()) %>%
.$count
))
Data is
structure(list(Id = 1:15, nos = c(4L, 2L, 3L, 2L, 3L, 2L, 2L,
3L, 1L, 5L, 2L, 1L, 2L, 1L, 1L), grade = c(3L, 3L, 3L, 3L, 3L,
2L, 2L, 4L, 4L, 3L, 3L, 4L, 3L, 3L, 4L), Date_Created = structure(c(16861,
16875, 16881, 16946, 17011, 17011, 17011, 17017, 17088, 17105,
17269, 17269, 17269, 17269, 17346), class = "Date"), location = structure(c(1L,
1L, 1L, 2L, 2L, 2L, 5L, 5L, 1L, 2L, 2L, 6L, 3L, 4L, 4L), .Label = c("Aus",
"Ind", "KA", "LA", "ML", "PA"), class = "factor")), row.names = c(NA,
-15L), class = "data.frame")
In your first image you are using summarise_() instead of summarise(), it is not the same.
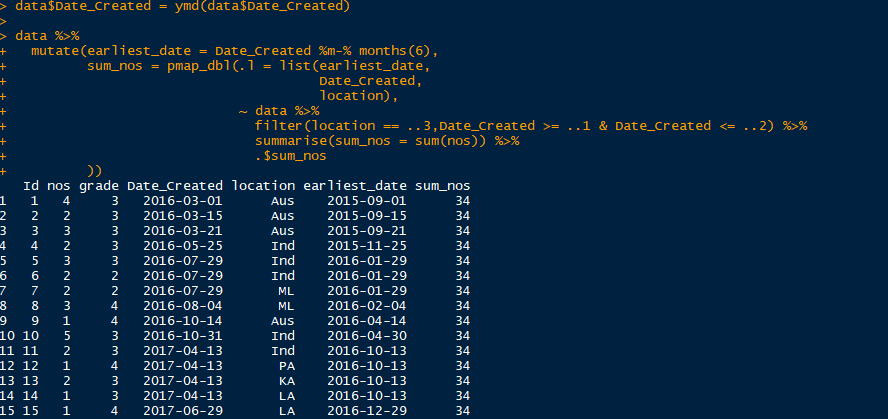
For your second image we don't know what the structure of data is, any chance you have grouped data? That could be causing the problem.
Also, I want to point out that you are not honoring your part of the deal, have you read the guides about how to use this forum? Please follow our guides when posting, using unformatted text and screenshots is considered a bad practice here.
Thank you. I have actually used the same data and the code which you shared. There is no change in the structure of the data.
I am sorry i am on deal and i tried to put it in the format which you shared. However it didn't work out. I will try again.
library(tidyverse)
library(lubridate)
library(plyr)
library(dplyr)
library(purrr)
library(magrittr)
data$Date_Created = ymd(data$Date_Created)
chk <- data %>%
mutate(earliest_date = Date_Created %m-% months(6),
count = pmap_dbl(.l = list(location,
grade,
earliest_date,
Date_Created),
~ data %>%
filter(location == ..1, grade == ..2, Date_Created >= ..3 & Date_Created <= ..4)%>%
summarise(count=n()) %>%
.$count
),
sum_nos = pmap_dbl(.l = list(location,
grade,
earliest_date,
Date_Created),
~ data %>%
filter(location == ..1, grade == ..2, Date_Created >= ..3 & Date_Created <= ..4) %>%
summarise(sum_nos = sum(nos)) %>%
.$sum_nos
)
)
structure(list(Id = 1:15, nos = c(4L, 2L, 3L, 2L, 3L, 2L, 2L,
3L, 1L, 5L, 2L, 1L, 2L, 1L, 1L), grade = c(3L, 3L, 3L, 3L, 3L,
2L, 2L, 4L, 4L, 3L, 3L, 4L, 3L, 3L, 4L), Date_Created = structure(c(16861,
16875, 16881, 16946, 17011, 17011, 17011, 17017, 17088, 17105,
17269, 17269, 17269, 17269, 17346), class = "Date"), location = structure(c(1L,
1L, 1L, 2L, 2L, 2L, 5L, 5L, 1L, 2L, 2L, 6L, 3L, 4L, 4L), .Label = c("Aus",
"Ind", "KA", "LA", "ML", "PA"), class = "factor")), row.names = c(NA,
-15L), class = "data.frame")
data %>%
mutate(earliest_date = Date_Created %m-% months(6),
count = pmap_dbl(.l = list(location,
grade,
earliest_date,
Date_Created),
~ data %>%
filter(location == ..1, grade == ..2, Date_Created >= ..3 & Date_Created <= ..4)%>%
summarise(count=n()) %>%
.$count
),
sum_nos = pmap_dbl(.l = list(location,
grade,
earliest_date,
Date_Created), ~ data %>%
filter(location == ..1, grade == ..2, Date_Created >= ..3 & Date_Created <= ..4) %>% summarise(sum_nos = sum(nos)) %>%
.$sum_nos )
)
Id nos grade Date_Created location earliest_date count sum_nos
1 1 4 3 2016-03-01 Aus 2015-09-01 15 34
2 2 2 3 2016-03-15 Aus 2015-09-15 15 34
3 3 3 3 2016-03-21 Aus 2015-09-21 15 34
4 4 2 3 2016-05-25 Ind 2015-11-25 15 34
5 5 3 3 2016-07-29 Ind 2016-01-29 15 34
6 6 2 2 2016-07-29 Ind 2016-01-29 15 34
7 7 2 2 2016-07-29 ML 2016-01-29 15 34
8 8 3 4 2016-08-04 ML 2016-02-04 15 34
9 9 1 4 2016-10-14 Aus 2016-04-14 15 34
10 10 5 3 2016-10-31 Ind 2016-04-30 15 34
11 11 2 3 2017-04-13 Ind 2016-10-13 15 34
12 12 1 4 2017-04-13 PA 2016-10-13 15 34
13 13 2 3 2017-04-13 KA 2016-10-13 15 34
14 14 1 3 2017-04-13 LA 2016-10-13 15 34
15 15 1 4 2017-06-29 LA 2016-12-29 15 34
What happens if you run the code inside reprex() function like this? do you get the same result?
reprex::reprex({
library(tidyverse)
library(lubridate)
data <- data.frame(
Id = c(1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L, 11L, 12L, 13L,
14L, 15L),
nos = c(4L, 2L, 3L, 2L, 3L, 2L, 2L, 3L, 1L, 5L, 2L, 1L, 2L, 1L, 1L),
grade = c(3L, 3L, 3L, 3L, 3L, 2L, 2L, 4L, 4L, 3L, 3L, 4L, 3L, 3L, 4L),
Date_Created = c("2016-03-01", "2016-03-15", "2016-03-21", "2016-05-25",
"2016-07-29", "2016-07-29", "2016-07-29", "2016-08-04",
"2016-10-14", "2016-10-31", "2017-04-13", "2017-04-13",
"2017-04-13", "2017-04-13", "2017-06-29"),
location = as.factor(c("Aus", "Aus", "Aus", "Ind", "Ind", "Ind", "ML",
"ML", "Aus", "Ind", "Ind", "PA", "KA", "LA",
"LA"))
)
data$Date_Created = ymd(data$Date_Created)
data %>%
mutate(earliest_date = Date_Created %m-% months(6),
count = pmap_dbl(.l = list(location,
grade,
earliest_date,
Date_Created),
~ data %>%
filter(location == ..1, grade == ..2, Date_Created >= ..3 & Date_Created <= ..4) %>%
summarise(count = n()) %>%
.$count
),
sum_nos = pmap_dbl(.l = list(location,
grade,
earliest_date,
Date_Created),
~ data %>%
filter(location == ..1, grade == ..2, Date_Created >= ..3 & Date_Created <= ..4) %>%
summarise(sum_nos = sum(nos)) %>%
.$sum_nos
)
)
})
Note: That would be the proper way of making a reproducible example, notice that it includes library calls, sample data and relevant code all in a self-contained reprex.
Thank you for the Note. Will do the same in future posts. When i run within reprex it's giving the following error
Error in file(con, "w") : cannot open the connection
In addition: Warning message:
In file(con, "w") : cannot open file 'C:/Users/P.R': Permission denied
That is very odd, I have never seen that kind of error when working with reprex, I recommend you to open a new topic about this issue, I suspect that it has something to do with non ANSI characters present in your windows user name.
Thanks. I have opened a new topic. But just out of curiosity i am not as expert in R but it should ideally run without reprex as well ? correct? is there a problem in version of R or any settings, or last time you mentioned conflicts. Please let me know thanks
i am using R.3.5.1 is it a problem?
It shouldn't be, this seems like a problem with your system setup some times R has problems with windows user names that have non ASCII characters, maybe that is your case.
I reinstalled R studio and updated the R package from R.3.5.1 To R.3.6.1 and it worked. Thank you so much for all the support. Really appreciate your time
This topic was automatically closed 7 days after the last reply. New replies are no longer allowed.