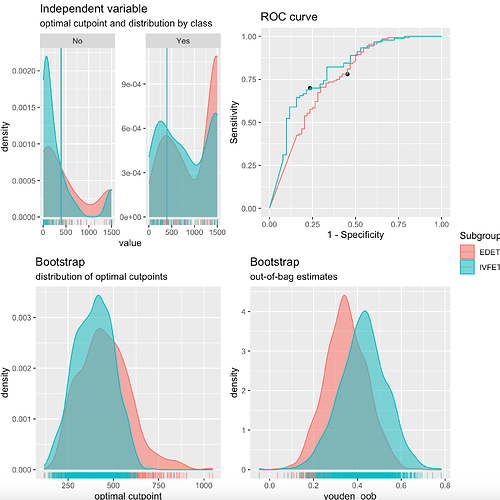
I am using the cutpointr package to generate cut off for a continuous variable. I work as prescribed but the plot objects generated are complex and a result of large gtable data. I want to format or edit the legend in the plot but I have failed totally with ggplot2
This is the code with cutpointr used to generate the cut off:
opt_cut_b_cycle.type<- cutpointr(hcgdf_v2, beta.hcg, livebirth.factor, cycle.type,
method = maximize_boot_metric,
metric = youden, boot_runs = 1000,
boot_stratify = TRUE,
na.rm = TRUE) %>% add_metric(list(ppv, npv, odds_ratio, risk_ratio, p_chisquared))
plot(opt_cut_b_cycle.type)
This is the plot generated:
- I want to edit the legend title from subgroup to Oocyte source
- I want to change the labels EDET to Donor, IVFET to Autologous
I tried working treating the plot object as a ggplot2 plot and running code such as, where p is the said plot object.
p + scale_fill_discrete(name = "Oocyte source", labels = c("Donor", "Autologous"))
Unfortunately, the console returns 'NULL'
This is an example data set:
hcgdf_v2 <-tibble(id = 1:10, beta.hcg = seq(from = 5, to = 1500, length.out = 10),
livebirth.factor = c("yes", "no", "yes", "no", "no", "yes", "yes", "no", "no", "yes"),
cycle.type = c("edet","ivfet","edet", "edet", "edet", "edet", "ivfet", "ivfet", "ivfet","edet"))
Thank you