The how part is straightforward, the what part needs closer examination.
# data
d <- data.frame(year = c(
2005, 2005, 2005, 2005, 2005, 2005,
2005, 2005, 2005, 2005, 2005, 2005, 2006, 2006, 2006,
2006, 2006, 2006, 2006, 2006, 2006, 2006, 2006, 2006,
2007, 2007, 2007, 2007, 2007, 2007, 2007, 2007, 2007,
2007, 2007, 2007, 2008, 2008, 2008, 2008, 2008, 2008,
2008, 2008, 2008, 2008, 2008, 2008, 2009, 2009, 2009,
2009, 2009, 2009, 2009, 2009, 2009, 2009, 2009, 2009,
2010, 2010, 2010, 2010, 2010, 2010, 2010, 2010, 2010,
2010, 2010, 2010, 2011, 2011, 2011, 2011, 2011, 2011,
2011, 2011, 2011, 2011, 2011, 2011, 2012, 2012, 2012,
2012, 2012, 2012, 2012, 2012, 2012, 2012, 2012, 2012,
2013, 2013, 2013, 2013, 2013, 2013, 2013, 2013, 2013,
2013, 2013, 2013, 2014, 2014, 2014, 2014, 2014, 2014,
2014, 2014, 2014, 2014, 2014, 2014
), month = c(
1, 2, 3,
4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10,
11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5,
6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12,
1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8,
9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3,
4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10,
11, 12
), h2 = c(
1, 1, 65, 1, 1, 54, 20, 1, 0, 1, 22, 38, 1, 2,
0, 1, 3, 29, 1, 1, 0, 55, 1, 3, 63, 1, 0, 4, 1, 4, 6, 2, 1, 0,
1, 1, 1, 2, 1, 3, 1, 1, 0, 3, 13, 49, 1, 2, 8, 3, 8, 4, 0, 0,
1, 1, 3, 1, 2, 0, 45, 1, 2, 13, 0, 1, 1, 1, 65, 49, 65, 63, 1,
8, 5, 20, 3, 52, 49, 1, 4, 1, 6, 1, 0, 72, 2, 1, 6, 1, 1, 94,
3, 3, 4, 49, 1, 1, 4, 1, 4, 0, 0, 0, 0, 2, 65, 3, 3, 1, 1, 78,
2, 0, 2, 1, 1, 1, 5, 1
), o18 = c(
55, 1, 25, 88, 65, 33, 1, 15,
11, 47, 23, 55, 1, 24, 31, 94, 23, 16, 78, 70, 33, 25, 99, 53,
71, 6, 1, 23, 25, 54, 71, 94, 73, 23, 97, 88, 14, 4, 91, 94,
25, 35, 40, 40, 25, 33, 47, 12, 23, 14, 33, 55, 55, 12, 40, 22,
31, 88, 36, 75, 55, 47, 46, 41, 31, 33, 34, 94, 97, 31, 14, 23,
18, 33, 16, 6, 16, 47, 34, 18, 47, 12, 47, 18, 33, 15, 77, 40,
49, 1, 75, 1, 51, 45, 94, 18, 12, 73, 33, 45, 23, 11, 35, 73,
71, 33, 47, 41, 41, 71, 35, 85, 16, 12, 31, 27, 39, 78, 49, 94
))
my_lm <- function(x) lm(h2 ~ o18,d[unlist(groups[x]),])
groups <- split(1:120,ceiling(seq_along(1:120) / 12))
# examine the last group, for example
my_lm(10) |> summary()
#>
#> Call:
#> lm(formula = h2 ~ o18, data = d[unlist(groups[x]), ])
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -22.596 -6.603 -2.540 1.229 57.466
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) -8.3904 12.6671 -0.662 0.523
#> o18 0.3403 0.2309 1.474 0.171
#>
#> Residual standard error: 20.99 on 10 degrees of freedom
#> Multiple R-squared: 0.1784, Adjusted R-squared: 0.09623
#> F-statistic: 2.171 on 1 and 10 DF, p-value: 0.1714
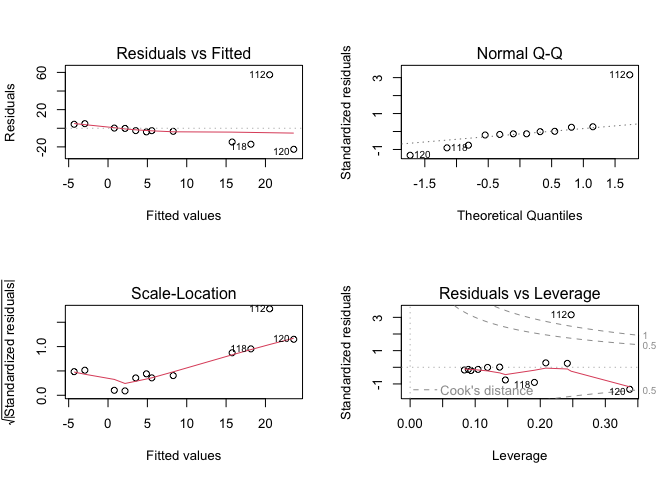
# show diagnostics
par(mfrow = c(2, 2))
plot(my_lm(10))
# use time series linear regression
library(fpp3)
#> ── Attaching packages ────────────────────────────────────────────── fpp3 0.5 ──
#> ✔ tibble 3.2.1 ✔ tsibble 1.1.3
#> ✔ dplyr 1.1.1 ✔ tsibbledata 0.4.1
#> ✔ tidyr 1.3.0 ✔ feasts 0.3.1
#> ✔ lubridate 1.9.2 ✔ fable 0.3.3
#> ✔ ggplot2 3.4.1 ✔ fabletools 0.3.2
#> ── Conflicts ───────────────────────────────────────────────── fpp3_conflicts ──
#> ✖ lubridate::date() masks base::date()
#> ✖ dplyr::filter() masks stats::filter()
#> ✖ tsibble::intersect() masks base::intersect()
#> ✖ tsibble::interval() masks lubridate::interval()
#> ✖ dplyr::lag() masks stats::lag()
#> ✖ tsibble::setdiff() masks base::setdiff()
#> ✖ tsibble::union() masks base::union()
# convert to time series in tsibble format
dts <- as_tsibble(ts(d[3:4], start = c(2005,1), frequency = 12))
dts <- dts |> pivot_wider(names_from = key)
dts |>
model(TSLM(h2 ~ o18)) |>
report()
#> Series: h2
#> Model: TSLM
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -13.897 -10.909 -8.484 -4.985 80.103
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 13.95972 3.57794 3.902 0.000159 ***
#> o18 -0.06263 0.07244 -0.865 0.388981
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Residual standard error: 21.38 on 118 degrees of freedom
#> Multiple R-squared: 0.006296, Adjusted R-squared: -0.002125
#> F-statistic: 0.7476 on 1 and 118 DF, p-value: 0.38898
Created on 2023-03-30 with reprex v2.0.2
