Let's start here. x and y differ in length()
Error in xy.coords(x, y, xlabel, ylabel, log) :
'x' and 'y' lengths differ
> x
[1] 15.5 22.5 29.5 36.5 43.5 50.5 57.5
> y
[1] 40 80 90 90
Compare the example, where dates and evaluation are of equal length
library(agricolae)
dates <- c(14, 21, 28) # days
# example 1: evaluation - vector
evaluation <- c(40, 80, 90)
audpc(evaluation, dates)
#> evaluation
#> 1015
# example 2: evaluation: dataframe nrow=1
evaluation <- data.frame(E1 = 40, E2 = 80, E3 = 90) # percentages
plot(dates, evaluation, type = "h", ylim = c(0, 100), col = "red", axes = FALSE)
title(cex.main = 0.8, main = "Absolute or Relative AUDPC\nTotal area = 100*(28-14)=1400")
lines(dates, evaluation, col = "red")
text(dates, evaluation + 5, evaluation)
text(18, 20, "A = (21-14)*(80+40)/2")
text(25, 60, "B = (28-21)*(90+80)/2")
text(25, 40, "audpc = A+B = 1015")
text(24.5, 33, "relative = audpc/area = 0.725")
abline(h = 0)
axis(1, dates)
axis(2, seq(0, 100, 5), las = 2)
lines(rbind(c(14, 40), c(14, 100)), lty = 8, col = "green")
lines(rbind(c(14, 100), c(28, 100)), lty = 8, col = "green")
lines(rbind(c(28, 90), c(28, 100)), lty = 8, col = "green")
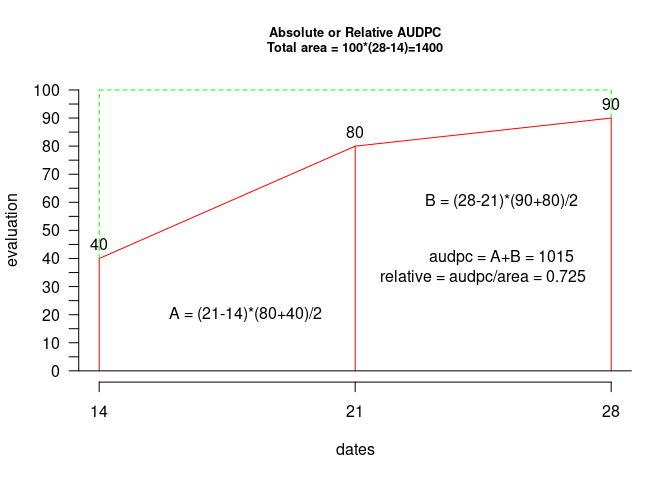
# It calculates audpc absolute
absolute <- audpc(evaluation, dates, type = "absolute")
print(absolute)
#> [1] 1015
rm(evaluation, dates, absolute)
# example 3: evaluation dataframe nrow>1
data(disease)
dates <- c(1, 2, 3) # week
evaluation <- disease[, c(4, 5, 6)]
# It calculates audpc relative
index <- audpc(evaluation, dates, type = "relative")
# Correlation between the yield and audpc
correlation(disease$yield, index, method = "kendall")
#>
#> Kendall's rank correlation tau
#>
#> data: disease$yield and index
#> z-norm = -3.326938 p-value = 0.0008780595
#> alternative hypothesis: true rho is not equal to 0
#> sample estimates:
#> tau
#> -0.5436832
# example 4: days infile
data(CIC)
comas <- CIC$comas
oxapampa <- CIC$oxapampa
dcomas <- names(comas)[9:16]
days <- as.numeric(substr(dcomas, 2, 3))
AUDPC <- audpc(comas[, 9:16], days)
relative <- audpc(comas[, 9:16], days, type = "relative")
h1 <- graph.freq(AUDPC, border = "red", density = 4, col = "blue")
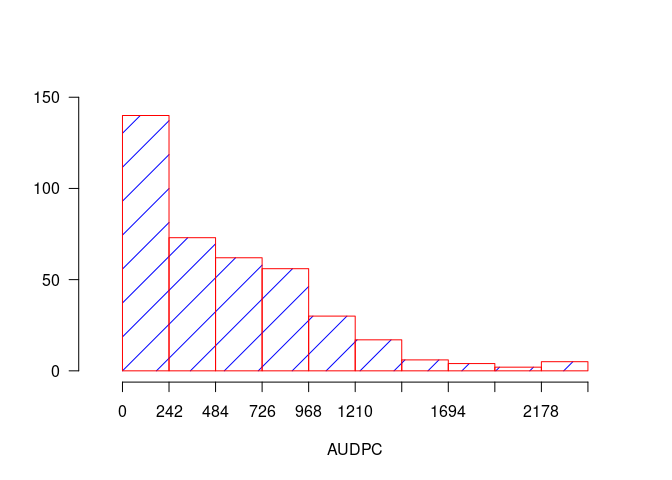
table.freq(h1)
h2 <- graph.freq(relative,
border = "red", density = 4, col = "blue",
frequency = 2, ylab = "relative frequency"
)
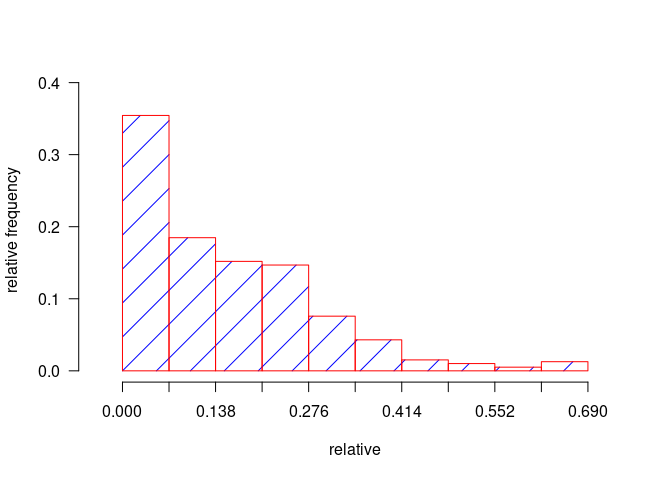
Created on 2022-11-22 by the reprex package (v2.0.1)


