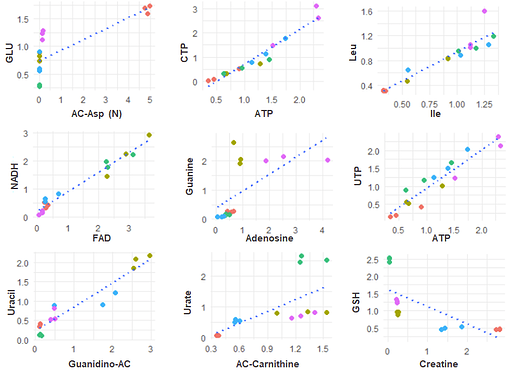
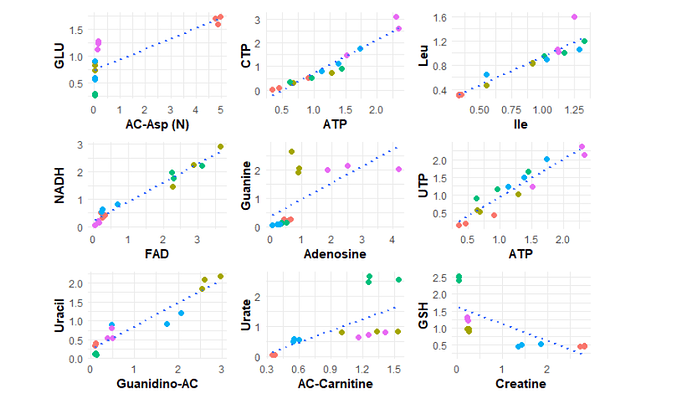
Is it possible to show a multiplet of plots in a grid-layot when the plots differ in their x- and y-axis (and these are not pairs)?
The first idea was of course facet_wrap() and actually it could be converted into a format that can be used, however then the axis aren't labelled.
Using the labeller function I can put the label into the title, however I want to have it on the axes as well.
See data and expected outcome below.
Is this in general possible with facet_wrap? Any ideas how to modify the axes?
Alternative? Run a loop, store the results in a list and use cowplot?
Demo = structure(list(row = c("AC-Asp (N)", "AC-Asp (N)", "AC-Asp (N)",
"AC-Asp (N)", "AC-Asp (N)", "AC-Asp (N)", "AC-Asp (N)", "AC-Asp (N)",
"AC-Asp (N)", "AC-Asp (N)", "AC-Asp (N)", "AC-Asp (N)", "AC-Asp (N)",
"AC-Asp (N)", "AC-Asp (N)", "AC-Carnitine", "AC-Carnitine", "AC-Carnitine",
"AC-Carnitine", "AC-Carnitine", "AC-Carnitine", "AC-Carnitine",
"AC-Carnitine", "AC-Carnitine", "AC-Carnitine", "AC-Carnitine",
"AC-Carnitine", "AC-Carnitine", "AC-Carnitine", "AC-Carnitine",
"Adenosine", "Adenosine", "Adenosine", "Adenosine", "Adenosine",
"Adenosine", "Adenosine", "Adenosine", "Adenosine", "Adenosine",
"Adenosine", "Adenosine", "Adenosine", "Adenosine", "Adenosine",
"ATP", "ATP", "ATP", "ATP", "ATP", "ATP", "ATP", "ATP", "ATP",
"ATP", "ATP", "ATP", "ATP", "ATP", "ATP", "ATP", "ATP", "ATP",
"ATP", "ATP", "ATP", "ATP", "ATP", "ATP", "ATP", "ATP", "ATP",
"ATP", "ATP", "ATP", "Creatine", "Creatine", "Creatine", "Creatine",
"Creatine", "Creatine", "Creatine", "Creatine", "Creatine", "Creatine",
"Creatine", "Creatine", "Creatine", "Creatine", "Creatine", "FAD",
"FAD", "FAD", "FAD", "FAD", "FAD", "FAD", "FAD", "FAD", "FAD",
"FAD", "FAD", "FAD", "FAD", "FAD", "Guanidino-AC", "Guanidino-AC",
"Guanidino-AC", "Guanidino-AC", "Guanidino-AC", "Guanidino-AC",
"Guanidino-AC", "Guanidino-AC", "Guanidino-AC", "Guanidino-AC",
"Guanidino-AC", "Guanidino-AC", "Guanidino-AC", "Guanidino-AC",
"Guanidino-AC", "Ile", "Ile", "Ile", "Ile", "Ile", "Ile", "Ile",
"Ile", "Ile", "Ile", "Ile", "Ile", "Ile", "Ile", "Ile"), Condition = c("Cond1",
"Cond1", "Cond1", "Cond2", "Cond2", "Cond2", "Cond3", "Cond3",
"Cond3", "Cond4", "Cond4", "Cond4", "Cond5", "Cond5", "Cond5",
"Cond1", "Cond1", "Cond1", "Cond2", "Cond2", "Cond2", "Cond3",
"Cond3", "Cond3", "Cond4", "Cond4", "Cond4", "Cond5", "Cond5",
"Cond5", "Cond1", "Cond1", "Cond1", "Cond2", "Cond2", "Cond2",
"Cond3", "Cond3", "Cond3", "Cond4", "Cond4", "Cond4", "Cond5",
"Cond5", "Cond5", "Cond1", "Cond1", "Cond1", "Cond1", "Cond1",
"Cond1", "Cond2", "Cond2", "Cond2", "Cond2", "Cond2", "Cond2",
"Cond3", "Cond3", "Cond3", "Cond3", "Cond3", "Cond3", "Cond4",
"Cond4", "Cond4", "Cond4", "Cond4", "Cond4", "Cond5", "Cond5",
"Cond5", "Cond5", "Cond5", "Cond5", "Cond1", "Cond1", "Cond1",
"Cond2", "Cond2", "Cond2", "Cond3", "Cond3", "Cond3", "Cond4",
"Cond4", "Cond4", "Cond5", "Cond5", "Cond5", "Cond1", "Cond1",
"Cond1", "Cond2", "Cond2", "Cond2", "Cond3", "Cond3", "Cond3",
"Cond4", "Cond4", "Cond4", "Cond5", "Cond5", "Cond5", "Cond1",
"Cond1", "Cond1", "Cond2", "Cond2", "Cond2", "Cond3", "Cond3",
"Cond3", "Cond4", "Cond4", "Cond4", "Cond5", "Cond5", "Cond5",
"Cond1", "Cond1", "Cond1", "Cond2", "Cond2", "Cond2", "Cond3",
"Cond3", "Cond3", "Cond4", "Cond4", "Cond4", "Cond5", "Cond5",
"Cond5"), Rep = c(1, 2, 3, 1, 2, 3, 1, 2, 3, 1, 2, 3, 1, 2, 3,
1, 2, 3, 1, 2, 3, 1, 2, 3, 1, 2, 3, 1, 2, 3, 1, 2, 3, 1, 2, 3,
1, 2, 3, 1, 2, 3, 1, 2, 3, 1, 1, 2, 2, 3, 3, 1, 1, 2, 2, 3, 3,
1, 1, 2, 2, 3, 3, 1, 1, 2, 2, 3, 3, 1, 1, 2, 2, 3, 3, 1, 2, 3,
1, 2, 3, 1, 2, 3, 1, 2, 3, 1, 2, 3, 1, 2, 3, 1, 2, 3, 1, 2, 3,
1, 2, 3, 1, 2, 3, 1, 2, 3, 1, 2, 3, 1, 2, 3, 1, 2, 3, 1, 2, 3,
1, 2, 3, 1, 2, 3, 1, 2, 3, 1, 2, 3, 1, 2, 3), RowValue = c(4.74478508369073,
4.84800266139503, 4.967034734681, 0.026615379200089, 0.0244870114269101,
0.0298224113724427, 0.0139647498371776, 0.0139238841993994, 0.0123895926726191,
0.0225933234667622, 0.0136490100262114, 0.027611846566471, 0.160374641580839,
0.131067996070496, 0.14854349984181, 0.374993004643239, 0.351665498230543,
0.359827276086599, 1.53056589424752, 1.32988946561918, 1.00110733389073,
1.26179208259026, 1.53642010883972, 1.2511884837821, 0.554221768119862,
0.548268022034505, 0.602478945280041, 1.25615871992003, 1.40908582976741,
1.15567684651475, 0.437633134872642, 0.584997375275487, 0.65295671963378,
0.90963029809557, 0.928900341924377, 0.675607239398359, 0.515055853028401,
0.352880652870411, 0.364195365245881, 0.296580321645923, 0.058959056336443,
0.210883586652197, 1.85673789697109, 4.18022306121108, 2.51313963323714,
0.451598537002974, 0.451598537002974, 0.905498537039748, 0.905498537039748,
0.347810869237147, 0.347810869237147, 1.28278033109806, 1.28278033109806,
0.63729113716726, 0.63729113716726, 0.679710782437389, 0.679710782437389,
1.44668532607411, 1.44668532607411, 0.621845490392747, 0.621845490392747,
0.959350504579393, 0.959350504579393, 1.12708177538952, 1.12708177538952,
1.73404732561652, 1.73404732561652, 1.38330942088472, 1.38330942088472,
2.33763074425223, 2.33763074425223, 2.29296722150237, 2.29296722150237,
1.51756643071633, 1.51756643071633, 2.81837374443456, 2.72194572304493,
2.8042564033653, 0.25726709435366, 0.260614572216056, 0.232976549590054,
0.0375646803368281, 0.0470845509019654, 0.0455050776375293, 1.35443825783268,
1.42718889466465, 1.85340483249492, 0.219562758244041, 0.225274122347207,
0.237410727289265, 0.268141045713833, 0.220780544393984, 0.333446723528368,
3.65054255388012, 2.8843544695329, 2.27263060529749, 3.10806726683728,
2.29852933980006, 2.24911722615092, 0.246650567349172, 0.694378896513481,
0.256060834424714, 0.0426520372425827, 0.148442375011389, 0.154211788187289,
0.126675847709749, 0.106257394696978, 0.112434326905105, 2.59384862809906,
2.9637262249831, 2.53332482949559, 0.0996217461640897, 0.11662261015213,
0.149457906078553, 0.480022874116761, 1.74512270402646, 2.07080503908742,
0.396700442977304, 0.4834266730824, 0.510269293170624, 0.32892836365911,
0.351068124623373, 0.328758406113227, 0.918395712897086, 0.914048141142347,
0.547157347496486, 1.33255433517194, 1.17203447268236, 1.01287858434647,
1.0295930315308, 1.2884334081136, 0.551150015644061, 1.11897250857575,
1.12285604344057, 1.2468746700899), column = c("GLU", "GLU",
"GLU", "GLU", "GLU", "GLU", "GLU", "GLU", "GLU", "GLU", "GLU",
"GLU", "GLU", "GLU", "GLU", "Urate", "Urate", "Urate", "Urate",
"Urate", "Urate", "Urate", "Urate", "Urate", "Urate", "Urate",
"Urate", "Urate", "Urate", "Urate", "Guanine", "Guanine", "Guanine",
"Guanine", "Guanine", "Guanine", "Guanine", "Guanine", "Guanine",
"Guanine", "Guanine", "Guanine", "Guanine", "Guanine", "Guanine",
"CTP", "UTP", "CTP", "UTP", "CTP", "UTP", "CTP", "UTP", "CTP",
"UTP", "CTP", "UTP", "CTP", "UTP", "CTP", "UTP", "CTP", "UTP",
"CTP", "UTP", "CTP", "UTP", "CTP", "UTP", "CTP", "UTP", "CTP",
"UTP", "CTP", "UTP", "GSH", "GSH", "GSH", "GSH", "GSH", "GSH",
"GSH", "GSH", "GSH", "GSH", "GSH", "GSH", "GSH", "GSH", "GSH",
"NADH", "NADH", "NADH", "NADH", "NADH", "NADH", "NADH", "NADH",
"NADH", "NADH", "NADH", "NADH", "NADH", "NADH", "NADH", "Uracil",
"Uracil", "Uracil", "Uracil", "Uracil", "Uracil", "Uracil", "Uracil",
"Uracil", "Uracil", "Uracil", "Uracil", "Uracil", "Uracil", "Uracil",
"Leu", "Leu", "Leu", "Leu", "Leu", "Leu", "Leu", "Leu", "Leu",
"Leu", "Leu", "Leu", "Leu", "Leu", "Leu"), Abs_Rank = c(1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 8L, 8L, 8L,
8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 5L, 5L, 5L, 5L,
5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 2L, 6L, 2L, 6L, 2L,
6L, 2L, 6L, 2L, 6L, 2L, 6L, 2L, 6L, 2L, 6L, 2L, 6L, 2L, 6L, 2L,
6L, 2L, 6L, 2L, 6L, 2L, 6L, 2L, 6L, 9L, 9L, 9L, 9L, 9L, 9L, 9L,
9L, 9L, 9L, 9L, 9L, 9L, 9L, 9L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L,
4L, 4L, 4L, 4L, 4L, 4L, 4L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L,
7L, 7L, 7L, 7L, 7L, 7L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L), ColValue = c(1.68905309798842, 1.59129707537898,
1.73014682816466, 0.738804909367395, 0.826681926608612, 0.895178856224703,
0.285155321657677, 0.274369883533457, 0.306301673125144, 0.593177459209097,
0.557915203523069, 0.908504723612642, 1.28192834653833, 1.12599021278152,
1.23386110382968, 0.0638736014216219, 0.0554392715639931, 0.059365505526676,
0.819553488649802, 0.83075617970686, 0.789708971424883, 2.65347341935594,
2.53289520765532, 2.45902532235367, 0.588758589805926, 0.488566738756838,
0.543133065163296, 0.707562235719935, 0.810894489554085, 0.632714882985723,
0.277654198359073, 0.240122595832372, 0.274000384916018, 1.90614143490259,
2.06385897103486, 2.63777498498008, 0.147113320510473, 0.132943615226974,
0.168307116815659, 0.106352060679067, 0.0750760221010915, 0.091421154973076,
2.0088936863467, 2.03114173073064, 2.14503107848986, 0.0860730968431118,
0.187997832715054, 0.528205779549168, 0.427593581420544, 0.0214807945521243,
0.1466770726327, 0.7287261963263, 1.01313930598596, 0.316888392426308,
0.571003825496993, 0.313779702248169, 0.532152108093237, 0.908665330358995,
1.66004104130761, 0.331656792262579, 0.905335782981719, 0.539234699480526,
1.16915693208703, 0.793117664219974, 1.24312996743378, 1.76397377006264,
2.02617017858924, 1.12905420920277, 1.50327111313035, 2.62388160033012,
2.13819896481415, 3.10956988937192, 2.38089161459363, 1.476938986499,
1.22793437517073, 0.458373052686694, 0.445942832624914, 0.480212080032847,
0.893169563353179, 0.969271950574871, 0.974733990968646, 2.50453665030777,
2.4161293829034, 2.52707200390318, 0.458928964576901, 0.498768343377741,
0.528496081767451, 1.32281567936353, 1.27942080850357, 1.23069145291835,
0.314685902391613, 0.50474977027588, 0.426474702632027, 2.92173061609381,
2.25896009532773, 1.45285587055888, 2.23321298736803, 1.76633141478557,
1.96539520489625, 0.532213053908337, 0.811957356342324, 0.636652164736579,
0.0580853068660453, 0.220637328547623, 0.134415507470217, 0.38873421451522,
0.345245451210446, 0.414986409500656, 2.09489876635595, 2.17559591013682,
1.85226006895812, 0.116513756488646, 0.135067696447721, 0.0986941693122155,
0.888429886024382, 0.909746838793847, 1.21250755548096, 0.530040191802449,
0.812105129504871, 0.531685966931552, 0.320881215916421, 0.307730576702745,
0.304837368835192, 0.844059484856992, 0.825840630868367, 0.464302642718788,
1.19379194848049, 1.00296336176796, 0.955020068460507, 0.889379355986826,
1.06132907840434, 0.649305759013481, 1.05868587583385, 1.0169409469303,
1.60020176187293), Combination = c("AC-Asp (N) & GLU", "AC-Asp (N) & GLU",
"AC-Asp (N) & GLU", "AC-Asp (N) & GLU", "AC-Asp (N) & GLU", "AC-Asp (N) & GLU",
"AC-Asp (N) & GLU", "AC-Asp (N) & GLU", "AC-Asp (N) & GLU", "AC-Asp (N) & GLU",
"AC-Asp (N) & GLU", "AC-Asp (N) & GLU", "AC-Asp (N) & GLU", "AC-Asp (N) & GLU",
"AC-Asp (N) & GLU", "AC-Carnitine & Urate", "AC-Carnitine & Urate",
"AC-Carnitine & Urate", "AC-Carnitine & Urate", "AC-Carnitine & Urate",
"AC-Carnitine & Urate", "AC-Carnitine & Urate", "AC-Carnitine & Urate",
"AC-Carnitine & Urate", "AC-Carnitine & Urate", "AC-Carnitine & Urate",
"AC-Carnitine & Urate", "AC-Carnitine & Urate", "AC-Carnitine & Urate",
"AC-Carnitine & Urate", "Adenosine & Guanine", "Adenosine & Guanine",
"Adenosine & Guanine", "Adenosine & Guanine", "Adenosine & Guanine",
"Adenosine & Guanine", "Adenosine & Guanine", "Adenosine & Guanine",
"Adenosine & Guanine", "Adenosine & Guanine", "Adenosine & Guanine",
"Adenosine & Guanine", "Adenosine & Guanine", "Adenosine & Guanine",
"Adenosine & Guanine", "ATP & CTP", "ATP & UTP", "ATP & CTP",
"ATP & UTP", "ATP & CTP", "ATP & UTP", "ATP & CTP", "ATP & UTP",
"ATP & CTP", "ATP & UTP", "ATP & CTP", "ATP & UTP", "ATP & CTP",
"ATP & UTP", "ATP & CTP", "ATP & UTP", "ATP & CTP", "ATP & UTP",
"ATP & CTP", "ATP & UTP", "ATP & CTP", "ATP & UTP", "ATP & CTP",
"ATP & UTP", "ATP & CTP", "ATP & UTP", "ATP & CTP", "ATP & UTP",
"ATP & CTP", "ATP & UTP", "Creatine & GSH", "Creatine & GSH",
"Creatine & GSH", "Creatine & GSH", "Creatine & GSH", "Creatine & GSH",
"Creatine & GSH", "Creatine & GSH", "Creatine & GSH", "Creatine & GSH",
"Creatine & GSH", "Creatine & GSH", "Creatine & GSH", "Creatine & GSH",
"Creatine & GSH", "FAD & NADH", "FAD & NADH", "FAD & NADH", "FAD & NADH",
"FAD & NADH", "FAD & NADH", "FAD & NADH", "FAD & NADH", "FAD & NADH",
"FAD & NADH", "FAD & NADH", "FAD & NADH", "FAD & NADH", "FAD & NADH",
"FAD & NADH", "Guanidino-AC & Uracil", "Guanidino-AC & Uracil",
"Guanidino-AC & Uracil", "Guanidino-AC & Uracil", "Guanidino-AC & Uracil",
"Guanidino-AC & Uracil", "Guanidino-AC & Uracil", "Guanidino-AC & Uracil",
"Guanidino-AC & Uracil", "Guanidino-AC & Uracil", "Guanidino-AC & Uracil",
"Guanidino-AC & Uracil", "Guanidino-AC & Uracil", "Guanidino-AC & Uracil",
"Guanidino-AC & Uracil", "Ile & Leu", "Ile & Leu", "Ile & Leu",
"Ile & Leu", "Ile & Leu", "Ile & Leu", "Ile & Leu", "Ile & Leu",
"Ile & Leu", "Ile & Leu", "Ile & Leu", "Ile & Leu", "Ile & Leu",
"Ile & Leu", "Ile & Leu")), class = c("grouped_df", "tbl_df",
"tbl", "data.frame"), row.names = c(NA, -135L), groups = structure(list(
row = c("AC-Asp (N)", "AC-Carnitine", "Adenosine", "ATP",
"Creatine", "FAD", "Guanidino-AC", "Ile"), .rows = structure(list(
1:15, 16:30, 31:45, 46:75, 76:90, 91:105, 106:120, 121:135), ptype = integer(0), class = c("vctrs_list_of",
"vctrs_vctr", "list"))), row.names = c(NA, -8L), class = c("tbl_df",
"tbl", "data.frame"), .drop = TRUE))
# Extract the labels for the titles
Titles = Demo %>%
distinct(Combination, Abs_Rank)
Plot_Titles = setNames(Titles$Combination, Titles$Abs_Rank)
# make the plot
ggplot(Demo,
aes(x = RowValue, y = ColValue)) +
geom_point(aes(colour = Condition),
size = 2, show.legend = FALSE) +
facet_wrap(~ Abs_Rank,
scales = "free",
labeller = as_labeller(Plot_Titles)) +
labs(aes(title = Combination)) +
theme_minimal() +
geom_smooth(method = "lm", se = FALSE,
linetype = "dotted")
expected outcome: