Hello,
I am struggling to use "Randomforest" analysis in R. Could you please review my code and give me some advice?
My dataset consists of two groups (HTP and OTP) and 36 metabolites.
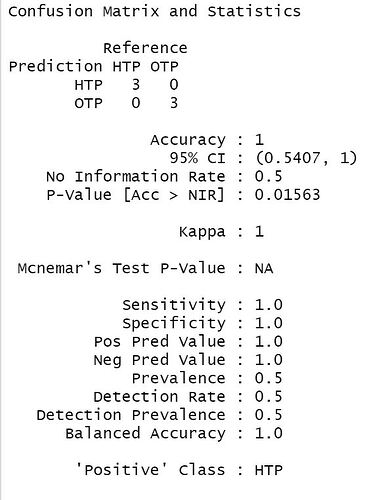
My concern is that the model is too accurate, and the results seem to be exaggerated. The accuracy of the confusion matrix and kappa value resulted in 1. Additionally, the VarImpPlot shows different results, which is why I used set.seed. As a first-time user of this package, please let me know if you find any mistakes.
Thank you in advance for your help.
Sorry for that "reprex" is not working on my R script.
library(randomForest)
library(caret)
library(ROCR)
library(tidyverse)
library(rfUtilities)
library(reprex)
RF <- tibble::tribble(
~Group, ~Acetate, ~Alanine, ~Aspartate, ~Benzoate, ~Butyrate, ~Carnitine, ~Creatine, ~Dimethyl.sulfone, ~Dimethylamine, ~Erythritol, ~Formate, ~Fumarate, ~Glucarate, ~Glycine, ~Glycylproline, ~Guanidoacetate, ~Isobutyrate, ~Isocitrate, ~Isoleucine, ~Isopropanol, ~Leucine, ~Methanol, ~Methionine, ~Methylsuccinate, ~N-Acetylaspartate, ~N-Methylhydantoin, ~Phenylacetate, ~Phenylalanine, ~Proline, ~Propionate, ~Pyroglutamate, ~Ribose, ~Syringate, ~Urea, ~Valine,
"OTP", 19490.9, 648.2, 266.6, 2.52, 7500.5, 1, 26.2, 1.8, 2.6, 99, 73.4, 42.8, 51, 91.8, 100, 13, 1384.6, 280.2, 255.8, 2.64, 278.8, 48.2, 123.2, 79.4, 22.6, 4.8, 639.8, 223.2, 480.8, 8939.1, 143.8, 952.6, 0.24, 24.88, 227.6,
"OTP", 33816.07, 264.2, 171.6, 2.52, 11174.5, 1, 5.2, 0.12, 2.2, 50.4, 54.4, 12.6, 2.68, 200, 59.6, 5.2, 740.8, 180.8, 109, 2.64, 151.2, 35.8, 52.6, 33.6, 19.6, 4.4, 533.4, 91, 219, 17597.8, 127.4, 104, 2.8, 24.88, 95.4,
"OTP", 22190.11, 609.2, 313.2, 2.52, 7980.7, 0.8, 0.32, 2, 1.4, 85.4, 55.4, 29.2, 2.68, 479.4, 116.6, 12.4, 1400, 124.8, 172.6, 2.64, 167.4, 15.8, 111.8, 40, 17.4, 0.24, 644, 5.12, 278.6, 10879, 24.44, 601, 6.8, 24.88, 340.4,
"OTP", 26250.8, 594, 584.4, 2.52, 10755.6, 8.2, 12.8, 0.12, 1.6, 60.8, 61.8, 17.2, 31.8, 621.6, 95.8, 16.4, 1506.8, 268.2, 272, 2.64, 347.8, 66.2, 152.4, 75, 27.6, 12.2, 835, 284, 622.4, 10728.2, 373, 476, 2.2, 24.88, 96.8,
"OTP", 25245.01, 57.6, 75.2, 2.52, 9995.9, 2.8, 17.4, 0.6, 1.2, 47.6, 50.6, 16.8, 2.68, 187.2, 65, 10.4, 1142, 85.8, 92.8, 16, 137, 33.6, 42.2, 14.8, 6, 1.6, 549.6, 73.4, 205, 12694.6, 24.44, 599.2, 1.6, 24.88, 65.8,
"OTP", 24234.8, 485.4, 220, 2.52, 9250.1, 0.6, 29, 0.6, 0.12, 54, 49.6, 22.8, 29.2, 284.6, 14.2, 93, 1065.4, 97.2, 159.8, 2.64, 196, 30.6, 71.4, 38.6, 3.4, 2.2, 557.2, 80.4, 299.4, 13446.6, 122.2, 289.6, 3.8, 24.88, 221.4,
"OTP", 23107.37, 527.4, 281, 2.52, 9068.6, 1.2, 2.8, 0.6, 6.8, 9.52, 60.8, 18.2, 24, 336.6, 7, 100.8, 1065, 113.2, 111.4, 2.64, 149.8, 35, 4.8, 66, 30.8, 1.2, 439, 104.4, 358.8, 11270.4, 145.6, 636, 5, 24.88, 239.8,
"OTP", 18462.74, 739, 292.8, 2.52, 6214.6, 5.2, 12.2, 1, 0.8, 102.2, 49.4, 19, 13.4, 521.8, 147.8, 39.2, 857.8, 190.6, 240.2, 2.64, 269.6, 33.8, 151.2, 134.2, 61.8, 0.24, 383, 197.6, 542.6, 8173.9, 321.6, 526.4, 13.4, 24.88, 338.2,
"OTP", 29897.34, 403.2, 291, 2.52, 9559.1, 0.8, 17.4, 0.12, 0.12, 49.6, 48, 8.4, 79, 30.2, 1.12, 7, 1020.8, 185.6, 164, 2.64, 215.4, 29.6, 79.8, 2.96, 25.2, 3.8, 504.6, 117.6, 321.8, 14609.3, 24.44, 442.6, 4.6, 24.88, 178,
"OTP", 32100.3, 47, 96.2, 2.52, 8477.4, 0.12, 22.6, 0.8, 3.6, 48.4, 5.56, 28, 97.4, 365.8, 11, 96, 1048.8, 137.4, 150.8, 2.64, 166.8, 3.6, 86.8, 77.6, 0.56, 2.4, 454.8, 149.4, 413, 17041, 202.8, 951, 6.6, 24.88, 197.4,
"HTP", 20776.8, 93.4, 72.6, 18.4, 3435.6, 0.12, 1.6, 7.4, 0.6, 9.52, 30, 12.8, 2.68, 16, 1.12, 37.8, 37.36, 10.08, 30, 28.4, 13.8, 12.8, 16.6, 2.96, 2.8, 0.24, 134.8, 5.12, 81.8, 4846.6, 24.44, 58, 0.24, 24.88, 30.6,
"HTP", 27524.4, 93, 80.2, 23.6, 4499.6, 0.12, 0.32, 0.12, 0.12, 9.52, 27.8, 0.72, 2.68, 62.2, 5.6, 1.8, 186.8, 79.8, 27.6, 28.6, 18, 6.4, 18, 2.96, 0.56, 0.24, 176, 5.12, 95.8, 6567, 24.44, 27.6, 0.24, 147.4, 5,
"HTP", 31638, 54.8, 68, 12.6, 4770, 0.12, 7.4, 7.4, 0.12, 9.52, 5.56, 0.72, 2.68, 61.6, 9.6, 8, 37.36, 10.08, 38, 13.2, 1.8, 13.4, 24.2, 2.96, 0.56, 1.6, 185, 25.6, 7.12, 6720.6, 24.44, 174.2, 0.24, 24.88, 0.92,
"HTP", 37312, 156, 105.4, 2.52, 7100.8, 0.6, 1.8, 12.4, 0.12, 9.52, 5.56, 8, 2.68, 3.6, 1.12, 2.4, 37.36, 10.08, 69, 37.8, 95.2, 16.8, 34.2, 2.96, 12.4, 0.24, 261.6, 59.8, 181, 8013, 24.44, 516, 10.2, 24.88, 0.92,
"HTP", 24217.6, 64.6, 11.36, 63.4, 4677.8, 0.12, 3, 5, 0.12, 9.52, 5.56, 5.6, 2.68, 39.2, 1.12, 2.6, 37.36, 10.08, 15.6, 45.6, 9, 13, 4.6, 2.96, 3.2, 0.24, 230.8, 5.12, 35.6, 5883, 24.44, 179.6, 1.2, 299, 0.92,
"HTP", 33942.4, 76, 56.8, 2.52, 5283, 0.12, 0.32, 2.8, 0.12, 9.52, 5.56, 7.6, 2.68, 44.4, 12.4, 1.6, 37.36, 10.08, 21.8, 31.8, 12.2, 0.72, 11.2, 2.96, 0.56, 0.24, 273.2, 5.12, 71.2, 8354.4, 24.44, 170.8, 0.24, 124.4, 4.6,
"HTP", 34943.2, 118.8, 87.4, 2.52, 6402, 0.12, 2.6, 3.4, 0.12, 9.52, 45.4, 9.8, 2.68, 73.8, 1.12, 0.32, 401.2, 51.4, 45.6, 34.8, 32.2, 9.6, 23.2, 2.96, 0.56, 0.24, 311.2, 5.12, 128.4, 8767.2, 24.44, 259, 0.24, 338.4, 21,
"HTP", 28161, 102, 11.36, 22.6, 4824.6, 0.12, 0.32, 8.8, 0.12, 9.52, 5.56, 6.4, 2.68, 61.4, 1.12, 6.8, 522.6, 10.08, 29.6, 28.8, 16.2, 4.4, 20.8, 2.96, 0.56, 2.2, 282.4, 5.12, 135.4, 4878.6, 24.44, 123, 0.24, 24.88, 11,
"HTP", 41891, 44.6, 11.36, 28.4, 6865.2, 0.12, 1.8, 1, 0.12, 47.8, 32.6, 0.72, 2.68, 47.4, 1.12, 0.32, 37.36, 112.8, 3.12, 21.4, 1.8, 3.8, 4.6, 2.96, 0.56, 0.24, 284.6, 5.12, 55, 10465.6, 24.44, 114, 0.24, 24.88, 11.6,
"HTP", 58430.8, 135.8, 11.36, 27.4, 9009.4, 5.4, 0.32, 2.2, 0.12, 52.2, 5.56, 3.6, 2.68, 54, 15, 7, 37.36, 50.4, 39.6, 39.6, 16.4, 0.72, 17.6, 2.96, 0.56, 0.24, 176.8, 5.12, 77.2, 16718, 24.44, 209.8, 0.24, 391.4, 18.2
)
RF$Group <- as.factor(RF$Group)
set.seed(123) # for reproducibility
trainIndex <- createDataPartition(RF$Group, p = 0.7, list = FALSE)
RF_train <- RF[trainIndex,]
RF_test <- RF[-trainIndex,]
ctrl <- trainControl(method = "cv", number = 99, savePredictions = TRUE)
set.seed(123)
RF_m_cv <- train(Group ~ ., data = RF_train, method = "rf",
trControl = ctrl, tuneLength = 10, ntree = 500)
set.seed(123)
RF_m <- randomForest(Group ~ ., data = RF_train,
ntree = 500, mtry = RF_m_cv$bestTune$mtry, importance = TRUE)
RF_m
pred <- predict(RF_m, RF_test)
cm <- confusionMatrix(pred, RF_test$Group)
cm
varImpPlot(RF_m)