Hi, I’m trying to use the function pivot_wider in order to have one column for Species and one column for each Plant Species.
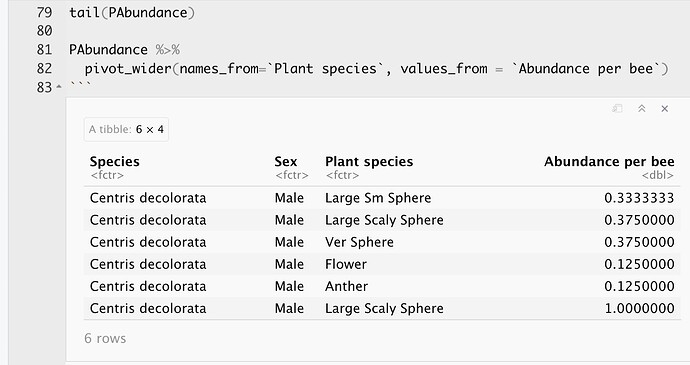
This is the data set:
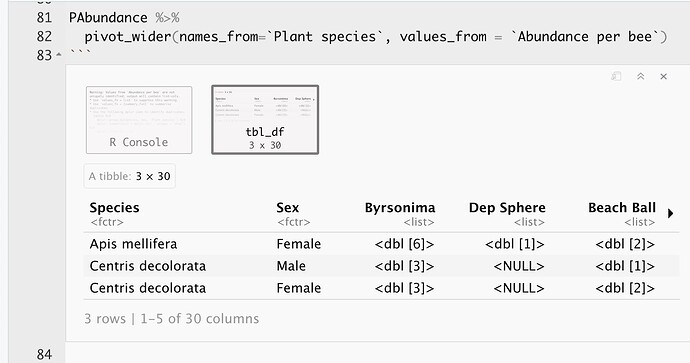
This is the result of using pivot_wider:
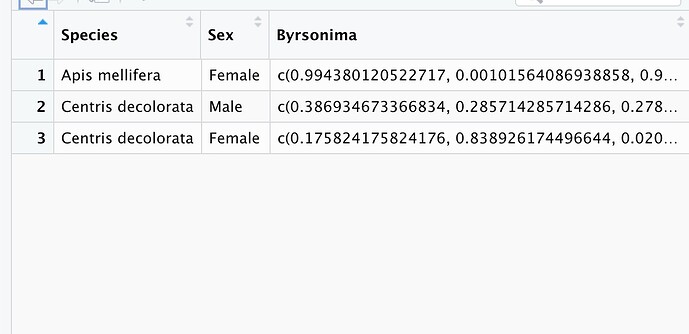
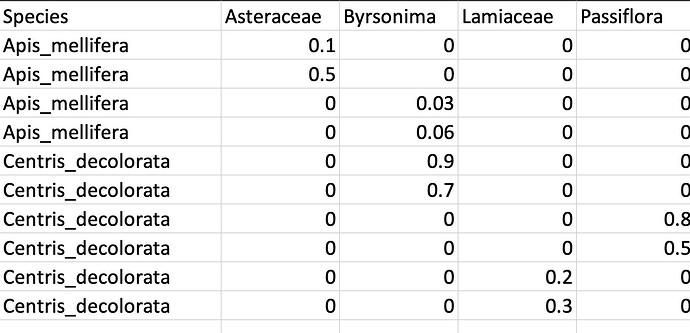
The observations are being grouped by each bee species and by Plant Species. I would like to ungroup the observations in order to see the multiple observations (rows) for each bee species. How can I obtain this using pivot_wider?
I tried using unlist() but it created multiple columns for each Plant Species; for example: Byrsonima1, Byrsonima2, etc.
Thank you!