Galal
May 27, 2023, 3:40pm
1
Hello everyone,
I'm using ggplot2 to create a correlation heatmap, I'm trying to add the p value as stars (***) inside the figure, this is the script that was used:
hp %>%
hp_filtered <-
hp_filtered %>%
I appreciate your help in advance
1 Like
Hi @Galal
suppressPackageStartupMessages(library(tidyverse))
library(Hmisc)
#>
#> Attaching package: 'Hmisc'
#> The following objects are masked from 'package:dplyr':
#>
#> src, summarize
#> The following objects are masked from 'package:base':
#>
#> format.pval, units
# Change one column name in TIME06 to get correct ordering
my_data<- read.delim(header = TRUE, sep=" ", text="
X E.coli L.salivarius B.fragilis B.longum TIME06 TIME12 TIME24
R1 0.008238399 0.037155326 0.005557831 0.023688994 1.776333 4.25300 16.39300
R2 0.001297156 0.090566197 0.007841161 0.012975925 8.140000 18.24925 25.48738
R3 0.002386935 0.062769327 0.024554017 0.015576090 5.125875 14.66462 25.98112
R4 0.002016223 0.006694006 0.008878337 0.007006086 7.150500 13.97425 16.58438
R5 0.003888820 0.001150177 0.008036232 0.003939090 3.910125 14.36300 18.49750
R6 0.015246245 0.000144155 0.012469594 0.008493679 3.214625 16.73150 33.40962
")
out <- rcorr(as.matrix(my_data[,2:8]), type = c("spearman"))
out$r
#> E.coli L.salivarius B.fragilis B.longum TIME06
#> E.coli 1.00000000 -0.7142857 0.08571429 0.08571429 -0.9428571
#> L.salivarius -0.71428571 1.0000000 -0.25714286 0.60000000 0.5428571
#> B.fragilis 0.08571429 -0.2571429 1.00000000 -0.20000000 0.1428571
#> B.longum 0.08571429 0.6000000 -0.20000000 1.00000000 -0.2571429
#> TIME06 -0.94285714 0.5428571 0.14285714 -0.25714286 1.0000000
#> TIME12 -0.25714286 0.2000000 0.31428571 -0.08571429 0.4857143
#> TIME24 0.14285714 -0.1428571 0.71428571 -0.08571429 0.1428571
#> TIME12 TIME24
#> E.coli -0.25714286 0.14285714
#> L.salivarius 0.20000000 -0.14285714
#> B.fragilis 0.31428571 0.71428571
#> B.longum -0.08571429 -0.08571429
#> TIME06 0.48571429 0.14285714
#> TIME12 1.00000000 0.82857143
#> TIME24 0.82857143 1.00000000
out$P
#> E.coli L.salivarius B.fragilis B.longum TIME06
#> E.coli NA 0.1107872 0.8717434 0.8717434 0.004804665
#> L.salivarius 0.110787172 NA 0.6227872 0.2080000 0.265702624
#> B.fragilis 0.871743440 0.6227872 NA 0.7040000 0.787172012
#> B.longum 0.871743440 0.2080000 0.7040000 NA 0.622787172
#> TIME06 0.004804665 0.2657026 0.7871720 0.6227872 NA
#> TIME12 0.622787172 0.7040000 0.5440933 0.8717434 0.328723032
#> TIME24 0.787172012 0.7871720 0.1107872 0.8717434 0.787172012
#> TIME12 TIME24
#> E.coli 0.62278717 0.78717201
#> L.salivarius 0.70400000 0.78717201
#> B.fragilis 0.54409329 0.11078717
#> B.longum 0.87174344 0.87174344
#> TIME06 0.32872303 0.78717201
#> TIME12 NA 0.04156268
#> TIME24 0.04156268 NA
# See: http://sthda.com/english/wiki/correlation-matrix-a-quick-start-guide-to-analyze-format-and-visualize-a-correlation-matrix-using-r-software
# ++++++++++++++++++++++++++++
# flattenCorrMatrix
# ++++++++++++++++++++++++++++
# cormat : matrix of the correlation coefficients
# pmat : matrix of the correlation p-values
flattenCorrMatrix <- function(cormat, pmat) {
ut <- upper.tri(cormat)
data.frame(
row = rownames(cormat)[row(cormat)[ut]],
column = rownames(cormat)[col(cormat)[ut]],
cor =(cormat)[ut],
p = pmat[ut]
)
}
hp <- flattenCorrMatrix(out$r, out$P)
hp_filtered <- hp %>%
filter(row %in% c("E.coli", "L.salivarius", "B.fragilis", "B.longum"),
column %in% c("TIME06", "TIME12", "TIME24"))
hp_filtered %>%
mutate(sig_text = case_when(
p >= 0.05 ~ "ns",
0.05 > p & p > 0.01 ~ "*",
0.01 >= p & p > 0.001 ~ "**",
p <=0.001 ~ "***",
.default = "error")) -> hp_filtered
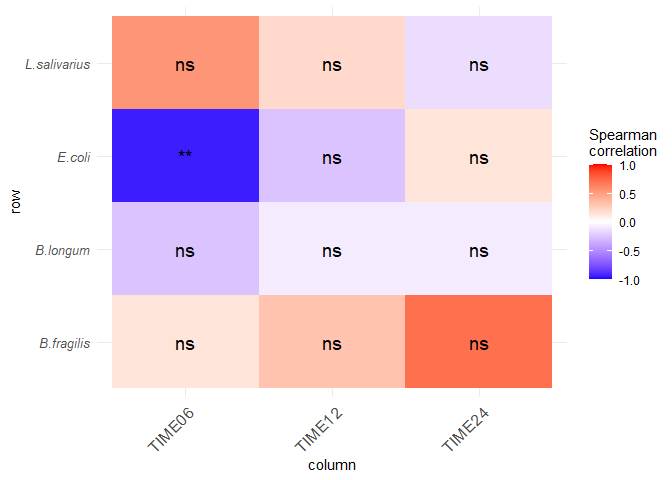
hp_filtered %>%
ggplot(aes(column, row, fill = cor)) +
geom_tile() +
scale_fill_gradient2(low = "blue", high = "red", mid = "white",
midpoint = 0, limit = c(-1,1), space = "Lab",
name="Spearman\ncorrelation") +
geom_text(label=hp_filtered$sig_text, size=5) +
theme_minimal() +
theme(axis.text.x = element_text(angle = 45, vjust = 1, size = 12, hjust = 1)) +
theme(axis.text.y = element_text(face ="italic", size = 10))
Created on 2023-05-28 with reprex v2.0.2
2 Likes
Galal
May 28, 2023, 1:35pm
3
Hi @DavoWW
system
June 4, 2023, 1:35pm
4
This topic was automatically closed 7 days after the last reply. New replies are no longer allowed.