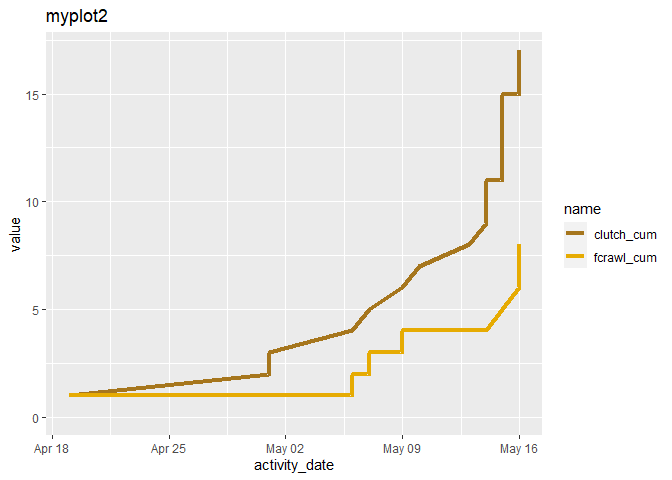
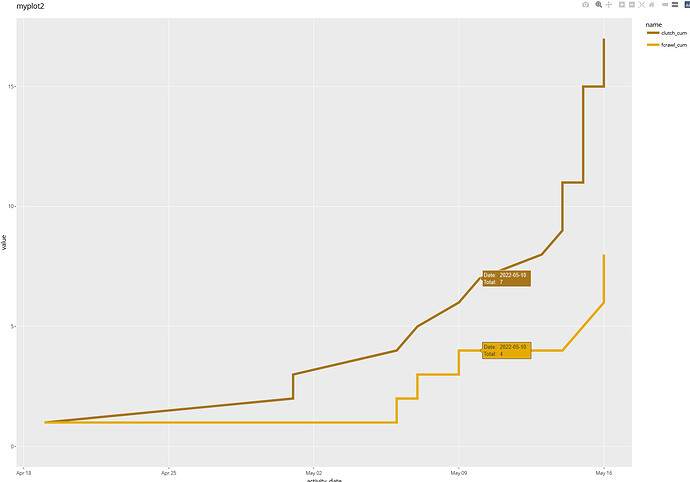
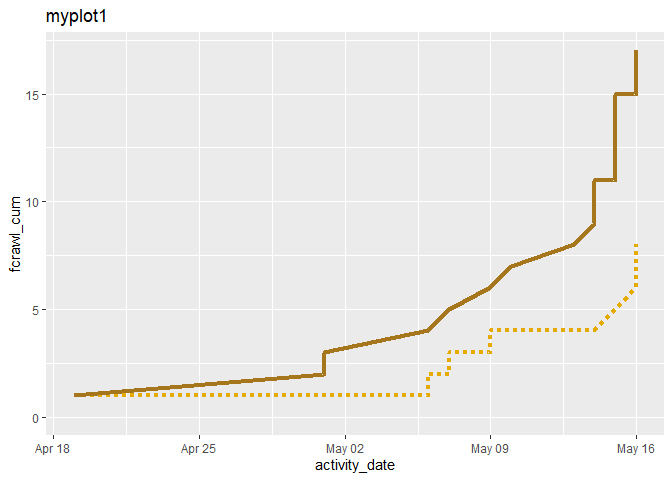
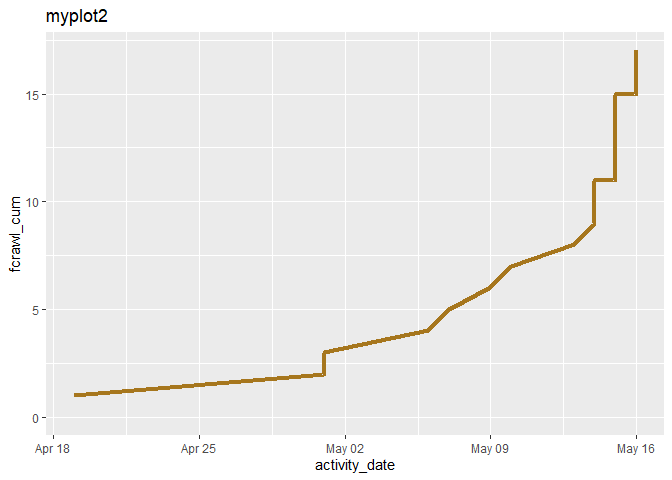
When I add a text line to a geom_line plot, that line disappears. Below I made two plots of two lines. In the second one, myplot2, the line, fcrawl_cum, has apparently disappeared.
``` r
library(tidyverse)
library("lubridate")
#>
#> Attaching package: 'lubridate'
#> The following objects are masked from 'package:base':
#>
#> date, intersect, setdiff, union
library(plotly)
#>
#> Attaching package: 'plotly'
#> The following object is masked from 'package:ggplot2':
#>
#> last_plot
#> The following object is masked from 'package:stats':
#>
#> filter
#> The following object is masked from 'package:graphics':
#>
#> layout
library("RColorBrewer")
library(htmlwidgets)
library("reprex")
activity <- c("N", "FB", "N", "N", "N", "FA", "N", "FA", "N", "FA", "N", "N", "N", "N", "N", "FA", "N", "N", "N", "N", "FA", "N", "N", "FA", "FA")
activity_date <- as.Date(c(NA, "2022-04-19", "2022-05-01", "2022-05-01", "2022-05-06", "2022-05-06", "2022-05-07", "2022-05-07", "2022-05-09", "2022-05-09", "2022-05-10", "2022-05-13", "2022-05-14", "2022-05-14", "2022-05-14", "2022-05-15", "2022-05-15", "2022-05-15", "2022-05-15", "2022-05-15", "2022-05-16", "2022-05-16", "2022-05-16", "2022-05-16", "2022-05-16"))
fcrawl_cum <- c(0L, 1L, 1L, 1L, 1L, 2L, 2L, 3L, 3L, 4L, 4L, 4L, 4L, 4L, 4L, 5L, 5L, 5L, 5L, 5L, 6L, 6L, 6L, 7L, 8L)
clutch_cum <- c(1L, 1L, 2L, 3L, 4L, 4L, 5L, 5L, 6L, 6L, 7L, 8L, 9L, 10L, 11L, 11L, 12L, 13L, 14L, 15L, 15L, 16L, 17L, 17L, 17L)
turtle_activity_gtm <- tibble(activity, activity_date, fcrawl_cum, clutch_cum)
the_pal <- RColorBrewer::brewer.pal(n = 8,"Dark2") #Set color palette.
myplot1 <-
ggplot() +
geom_line(data = turtle_activity_gtm,
aes(x=activity_date, y=fcrawl_cum),
na.rm = TRUE,
linetype = "111111",
linewidth = 1.5, color = the_pal[6]) +
geom_line(data = turtle_activity_gtm,
aes(x=activity_date, y=clutch_cum),
na.rm = TRUE,
linewidth = 1.5,
color = the_pal[7]) +
labs(title = "myplot1")
myplot1
myplot2 <-
ggplot() +
geom_line(data = turtle_activity_gtm,
aes(x=activity_date, y=fcrawl_cum,
text = paste("Date: ", as.Date(activity_date),
"<br>Total: ", fcrawl_cum)),
na.rm = TRUE,
linetype = "111111",
linewidth = 1.5, color = the_pal[6]) +
geom_line(data = turtle_activity_gtm,
aes(x=activity_date, y=clutch_cum),
na.rm = TRUE,
linewidth = 1.5,
color = the_pal[7]) +
labs(title = "myplot2")
#> Warning in geom_line(data = turtle_activity_gtm, aes(x = activity_date, :
#> Ignoring unknown aesthetics: text
myplot2
Created on 2023-02-05 with reprex v2.0.2
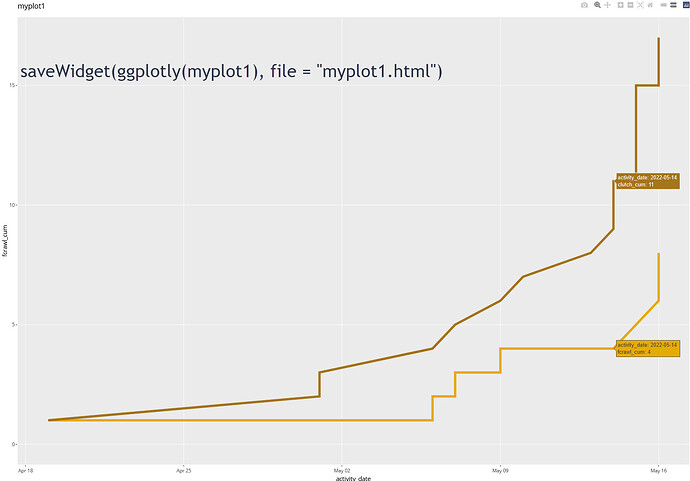
If I use
ggplotly(myplot1)
See image in reply.
the data points appear as expected although not in a format I like.
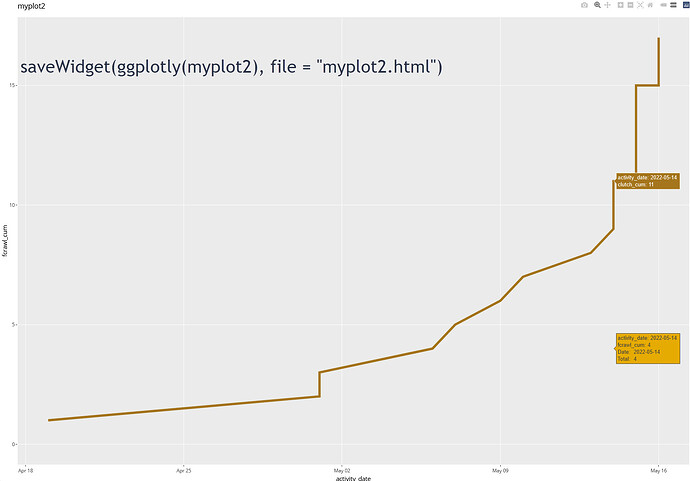
If I use
ggplotly(myplot2)
See image in reply.
the line with the text line added is still not there. However, the data points still appear for missing line.
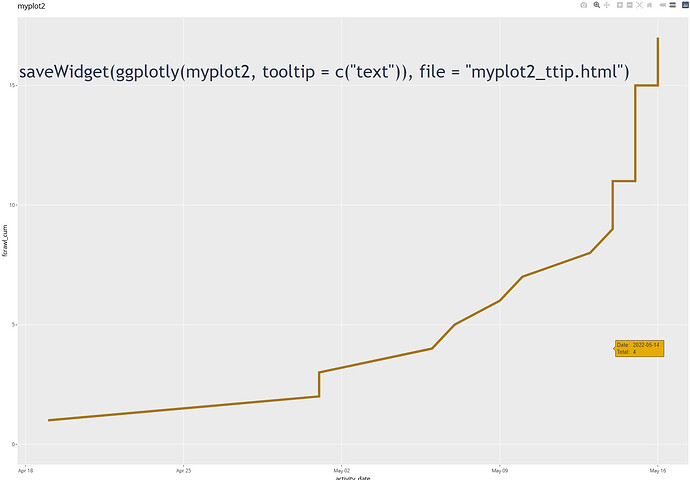
If I use ggplotly with the added tooltip,
ggplotly(myplot2, tooltip = c("text"))
See image in reply.
the label is missing for the line without the added text line but the label is exactly as written in the text line.
How can I do this properly so that both lines show with the added tooltip? I eventually want both lines to have their own text lines added. This is a very simplified chart. One I can get past this problem, I plan to eventually add a lot more items to this chart with a full data set.
Thanks,
Jeff